You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000025_00301
You are here: Home > Sequence: MGYG000000025_00301
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Staphylococcus nepalensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Staphylococcales; Staphylococcaceae; Staphylococcus; Staphylococcus nepalensis | |||||||||||
| CAZyme ID | MGYG000000025_00301 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | putative transglycosylase IsaA | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 333731; End: 334471 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PLN03198 | PLN03198 | 3.74e-04 | 27 | 178 | 9 | 155 | delta6-acyl-lipid desaturase; Provisional |
| cd00254 | LT-like | 0.006 | 182 | 217 | 6 | 45 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATH59179.1 | 2.37e-165 | 1 | 246 | 1 | 246 |
| ATH64271.1 | 2.37e-165 | 1 | 246 | 1 | 246 |
| AWI43631.1 | 2.37e-165 | 1 | 246 | 1 | 246 |
| AYX90777.1 | 4.26e-136 | 1 | 246 | 1 | 246 |
| VDZ21629.1 | 5.12e-126 | 1 | 246 | 1 | 248 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7IY64 | 1.28e-114 | 1 | 246 | 1 | 243 | Probable transglycosylase IsaA OS=Staphylococcus xylosus OX=1288 GN=isaA PE=1 SV=1 |
| Q4L980 | 1.77e-77 | 1 | 246 | 1 | 238 | Probable transglycosylase IsaA OS=Staphylococcus haemolyticus (strain JCSC1435) OX=279808 GN=isaA PE=3 SV=1 |
| Q4A0G5 | 7.66e-54 | 1 | 205 | 1 | 209 | Probable transglycosylase IsaA OS=Staphylococcus saprophyticus subsp. saprophyticus (strain ATCC 15305 / DSM 20229 / NCIMB 8711 / NCTC 7292 / S-41) OX=342451 GN=isaA PE=3 SV=1 |
| Q2YWD9 | 3.47e-37 | 1 | 230 | 1 | 223 | Probable transglycosylase IsaA OS=Staphylococcus aureus (strain bovine RF122 / ET3-1) OX=273036 GN=isaA PE=3 SV=1 |
| Q2FV52 | 9.69e-37 | 1 | 230 | 1 | 223 | Probable transglycosylase IsaA OS=Staphylococcus aureus (strain NCTC 8325 / PS 47) OX=93061 GN=isaA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
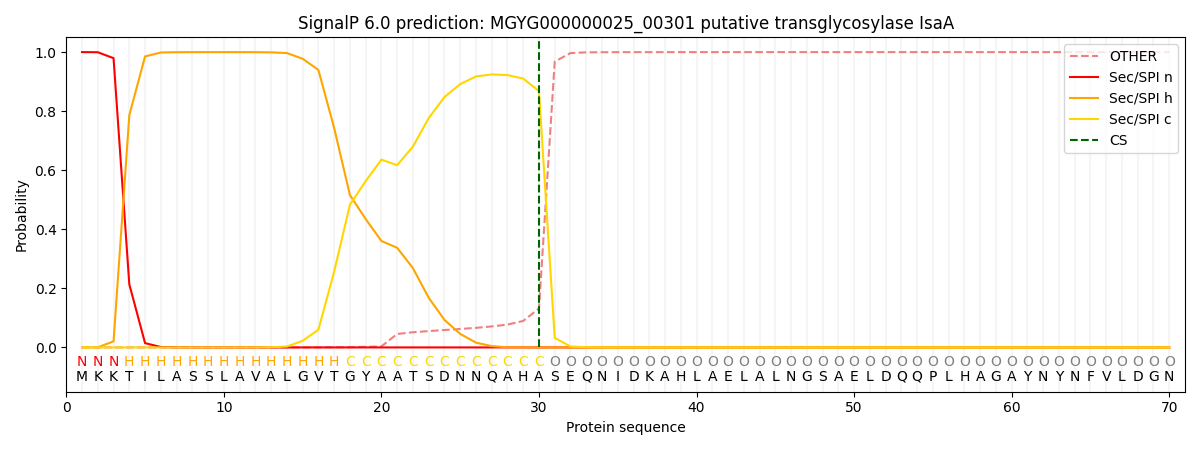
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000558 | 0.998430 | 0.000246 | 0.000289 | 0.000235 | 0.000205 |
