You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000029_00158
You are here: Home > Sequence: MGYG000000029_00158
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
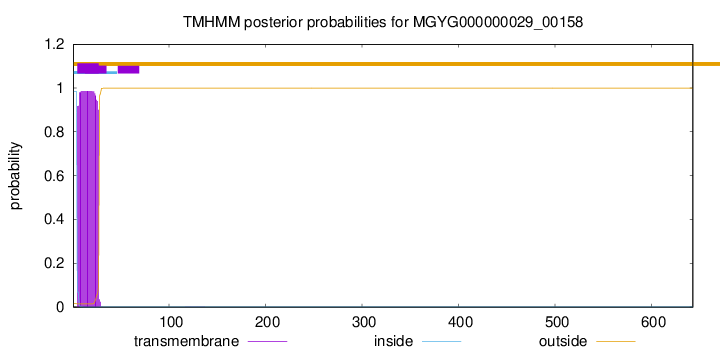
TMHMM annotations
Basic Information help
| Species | Bacteroides finegoldii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides finegoldii | |||||||||||
| CAZyme ID | MGYG000000029_00158 | |||||||||||
| CAZy Family | GH146 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 209199; End: 211130 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH146 | 52 | 557 | 5.6e-201 | 0.9980119284294234 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07944 | Glyco_hydro_127 | 0.0 | 52 | 557 | 1 | 503 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 1.79e-124 | 50 | 593 | 9 | 521 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT82455.1 | 0.0 | 1 | 643 | 1 | 643 |
| QUT22810.1 | 0.0 | 1 | 643 | 1 | 643 |
| QRM97805.1 | 0.0 | 1 | 643 | 1 | 643 |
| QGT69489.1 | 0.0 | 1 | 643 | 1 | 643 |
| QDM08600.1 | 0.0 | 1 | 643 | 1 | 643 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6YQH_AAA | 1.35e-118 | 44 | 643 | 24 | 653 | ChainAAA, Acetyl-CoA carboxylase, biotin carboxylase [Bacteroides thetaiotaomicron VPI-5482] |
| 5OPJ_A | 1.92e-113 | 44 | 643 | 24 | 653 | Beta-L-arabinofuranosidase[Bacteroides thetaiotaomicron] |
| 4QJY_A | 1.32e-18 | 244 | 564 | 201 | 567 | Crystalstructure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QJY_B Crystal structure of native Ara127N, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 4QK0_A | 9.36e-18 | 244 | 564 | 201 | 567 | Crystalstructure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_B Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_C Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus],4QK0_D Crystal structure of Ara127N-Se, a GH127 beta-L-arabinofuranosidase from Geobacillus Stearothermophilus T6 [Geobacillus stearothermophilus] |
| 5MQO_A | 8.74e-16 | 347 | 555 | 381 | 605 | Glycosidehydrolase BT_1003 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
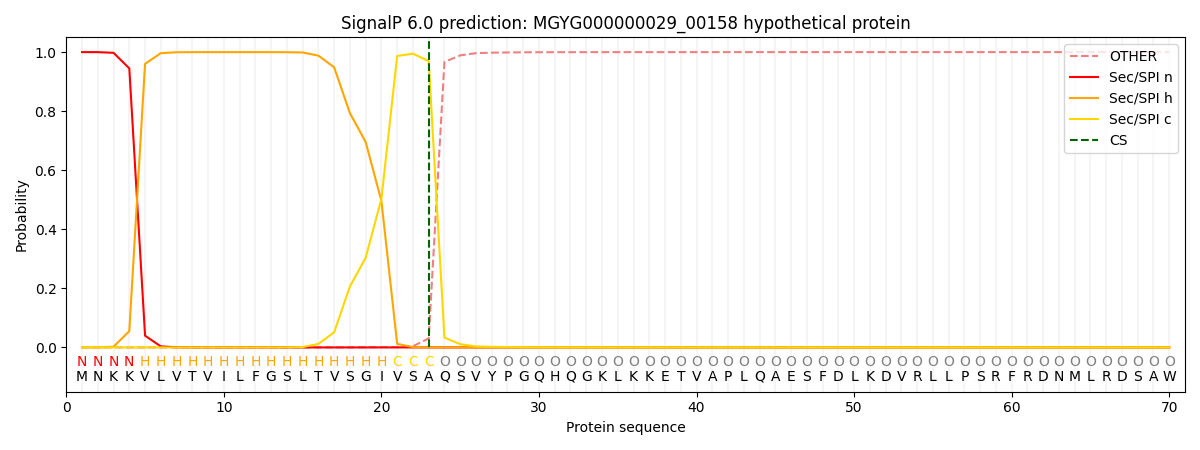
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000422 | 0.998835 | 0.000230 | 0.000175 | 0.000162 | 0.000148 |