You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000029_00848
You are here: Home > Sequence: MGYG000000029_00848
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides finegoldii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides finegoldii | |||||||||||
| CAZyme ID | MGYG000000029_00848 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 31211; End: 32557 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 326 | 443 | 2.5e-17 | 0.8870967741935484 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08522 | DUF1735 | 2.70e-14 | 174 | 288 | 1 | 120 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| pfam00754 | F5_F8_type_C | 3.55e-13 | 324 | 441 | 6 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam08522 | DUF1735 | 4.51e-09 | 38 | 159 | 7 | 120 | Domain of unknown function (DUF1735). This domain of unknown function is found in a number of bacterial proteins including acylhydrolases. The structure of this domain has a beta-sandwich fold. |
| cd00057 | FA58C | 5.03e-05 | 337 | 444 | 30 | 143 | Substituted updates: Jan 31, 2002 |
| cd08366 | APC10 | 0.002 | 331 | 374 | 19 | 57 | APC10 subunit of the anaphase-promoting complex (APC) that mediates substrate ubiquitination. This model represents the single domain protein APC10, a subunit of the anaphase-promoting complex (APC), which is a multi-subunit E3 ubiquitin ligase. E3 ubiquitin ligases mediate substrate ubiquitination (or ubiquitylation), a vital component of the ubiquitin-26S proteasome pathway for selective proteolytic degradation. The APC (also known as the cyclosome), is a cell cycle-regulated E3 ubiquitin ligase that controls important transitions in mitosis and the G1 phase by ubiquitinating regulatory proteins, thereby targeting them for degradation. In mitosis, the APC initiates sister chromatid separation by ubiquitinating the anaphase inhibitor securin and triggers exit from mitosis by ubiquitinating cyclin B. The C-terminus of APC10 binds to CDC27/APC3, an APC subunit that contains multiple tetratrico peptide repeats. APC10 domains are homologous to the DOC1 domains present in the HECT (Homologous to the E6-AP Carboxyl Terminus) E3 ubiquitin ligase protein, and the Cullin-RING (Really Interesting New Gene) E3 ubiquitin ligase complex. The APC10/DOC1 domain forms a beta-sandwich structure that is related in architecture to the galactose-binding domain-like fold; their sequences are quite dissimilar, however, and are not included here. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT72760.1 | 6.30e-281 | 1 | 448 | 1 | 449 |
| QUT88787.1 | 1.85e-183 | 9 | 448 | 7 | 447 |
| BCA52367.1 | 6.92e-176 | 9 | 447 | 8 | 446 |
| QUT73146.1 | 6.92e-176 | 9 | 447 | 8 | 446 |
| QUT40999.1 | 5.63e-175 | 9 | 447 | 8 | 446 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3F2Z_A | 3.87e-25 | 304 | 442 | 2 | 143 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
| 2KD7_A | 4.10e-21 | 303 | 442 | 1 | 140 | ChainA, Putative chitobiase [Bacteroides thetaiotaomicron VPI-5482] |
| 3GGL_A | 2.56e-20 | 304 | 442 | 12 | 150 | X-RayStructure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. [Bacteroides thetaiotaomicron],6OE2_A X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. Re-refinement of 3GGL with correct metal Mn replacing Zn. New metal confirmed with PIXE analysis of original sample. [Bacteroides thetaiotaomicron] |
| 2J7M_A | 8.81e-07 | 316 | 446 | 15 | 149 | Characterizationof a Family 32 CBM [Clostridium perfringens] |
| 2J1A_A | 8.97e-07 | 316 | 446 | 16 | 150 | Structureof CBM32 from Clostridium perfringens beta-N- acetylhexosaminidase GH84C in complex with galactose [Clostridium perfringens ATCC 13124],2J1E_A High Resolution Crystal Structure of CBM32 from a N-acetyl-beta- hexosaminidase in complex with lacNAc [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
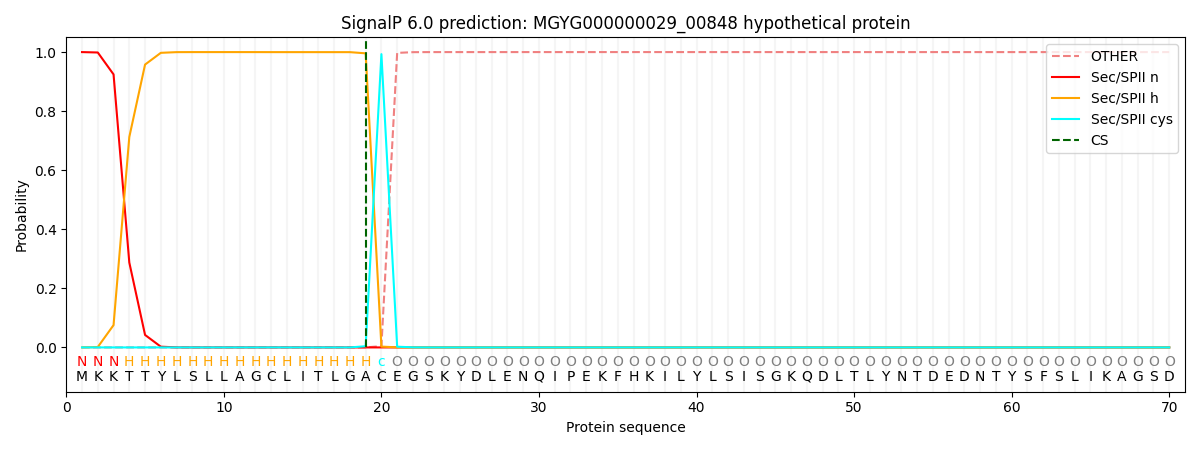
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000041 | 0.000000 | 0.000000 | 0.000000 |
