You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000032_03650
You are here: Home > Sequence: MGYG000000032_03650
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
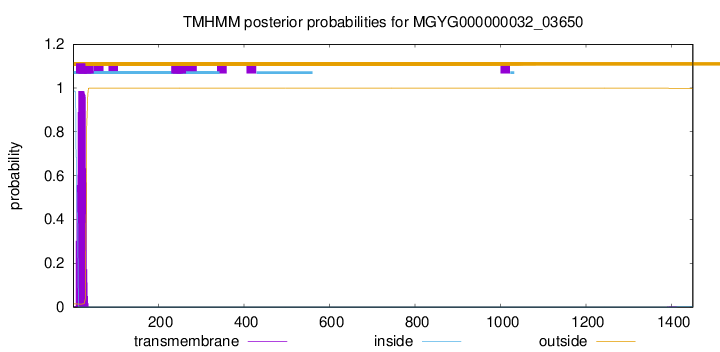
TMHMM annotations
Basic Information help
| Species | Hungatella effluvii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Hungatella; Hungatella effluvii | |||||||||||
| CAZyme ID | MGYG000000032_03650 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 117868; End: 122220 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 644 | 1205 | 1.9e-162 | 0.9938900203665988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5263 | COG5263 | 7.18e-18 | 1347 | 1446 | 222 | 313 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| NF033838 | PspC_subgroup_1 | 5.22e-17 | 1289 | 1447 | 422 | 586 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
| NF033840 | PspC_relate_1 | 2.00e-16 | 1307 | 1445 | 472 | 599 | PspC-related protein choline-binding protein 1. Members of this family share C-terminal homology to the choline-binding form of the pneumococcal surface antigen PspC, but not to its allelic LPXTG-anchored forms because they lack the choline-binding repeat region. Members of this family should not be confused with PspC itself, whose identity and function reflect regions N-terminal to the choline-binding region. See Iannelli, et al. (PMID: 11891047) for information about the different allelic forms of PspC. |
| COG5263 | COG5263 | 1.35e-13 | 1299 | 1449 | 93 | 257 | Glucan-binding domain (YG repeat) [Carbohydrate transport and metabolism]. |
| NF033838 | PspC_subgroup_1 | 4.36e-13 | 1289 | 1446 | 435 | 683 | pneumococcal surface protein PspC, choline-binding form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. The other form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AIY03209.1 | 7.47e-299 | 84 | 1211 | 26 | 1168 |
| SDS97815.1 | 1.11e-297 | 90 | 1212 | 51 | 1191 |
| QLL50162.1 | 6.06e-210 | 254 | 1205 | 33 | 977 |
| QLL52630.1 | 6.06e-210 | 254 | 1205 | 33 | 977 |
| ALA27928.1 | 6.06e-210 | 254 | 1205 | 33 | 977 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 5.54e-61 | 645 | 1205 | 10 | 576 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 5GQC_A | 7.73e-55 | 639 | 1137 | 13 | 571 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 7.04e-47 | 645 | 1137 | 15 | 583 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
| 6KQT_A | 2.00e-42 | 646 | 1019 | 247 | 644 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - native protein [Eubacterium ramulus ATCC 29099] |
| 6KQS_A | 4.78e-42 | 646 | 1019 | 247 | 644 | CrystalStructure of GH136 lacto-N-biosidase from Eubacterium ramulus - selenomethionine derivative [Eubacterium ramulus ATCC 29099] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
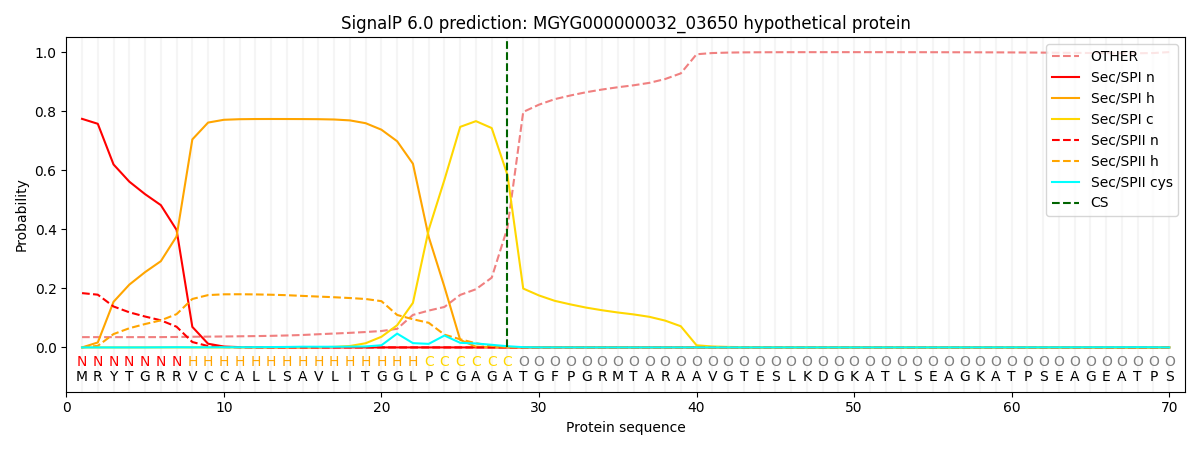
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.040357 | 0.762281 | 0.187648 | 0.008191 | 0.001145 | 0.000355 |