You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000033_00677
You are here: Home > Sequence: MGYG000000033_00677
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-41 sp900066215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; CAG-41; CAG-41 sp900066215 | |||||||||||
| CAZyme ID | MGYG000000033_00677 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52816; End: 54225 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.67e-81 | 227 | 460 | 14 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 2.43e-29 | 304 | 447 | 3 | 142 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam02368 | Big_2 | 2.02e-09 | 141 | 218 | 2 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam01832 | Glucosaminidase | 1.71e-07 | 317 | 392 | 4 | 76 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| COG5492 | YjdB | 7.05e-06 | 158 | 221 | 198 | 262 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACL75914.1 | 9.12e-74 | 238 | 460 | 319 | 544 |
| QEL36502.1 | 2.09e-73 | 238 | 460 | 677 | 902 |
| BBI95942.1 | 3.91e-73 | 238 | 460 | 675 | 900 |
| AOP96031.1 | 1.04e-72 | 238 | 460 | 675 | 900 |
| AHG23324.1 | 1.29e-72 | 240 | 461 | 212 | 435 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXP_A | 7.81e-49 | 282 | 465 | 69 | 250 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 2.25e-48 | 283 | 460 | 61 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 2.57e-48 | 282 | 465 | 99 | 280 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 4PI7_A | 9.21e-44 | 285 | 448 | 55 | 215 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 6.85e-43 | 285 | 448 | 55 | 215 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 1.30e-48 | 263 | 460 | 682 | 879 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q8NX96 | 1.58e-43 | 283 | 460 | 1073 | 1255 | Bifunctional autolysin OS=Staphylococcus aureus (strain MW2) OX=196620 GN=atl PE=3 SV=1 |
| Q6GAG0 | 1.58e-43 | 283 | 460 | 1067 | 1249 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q931U5 | 2.13e-43 | 283 | 460 | 1065 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| Q99V41 | 2.13e-43 | 283 | 460 | 1065 | 1247 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
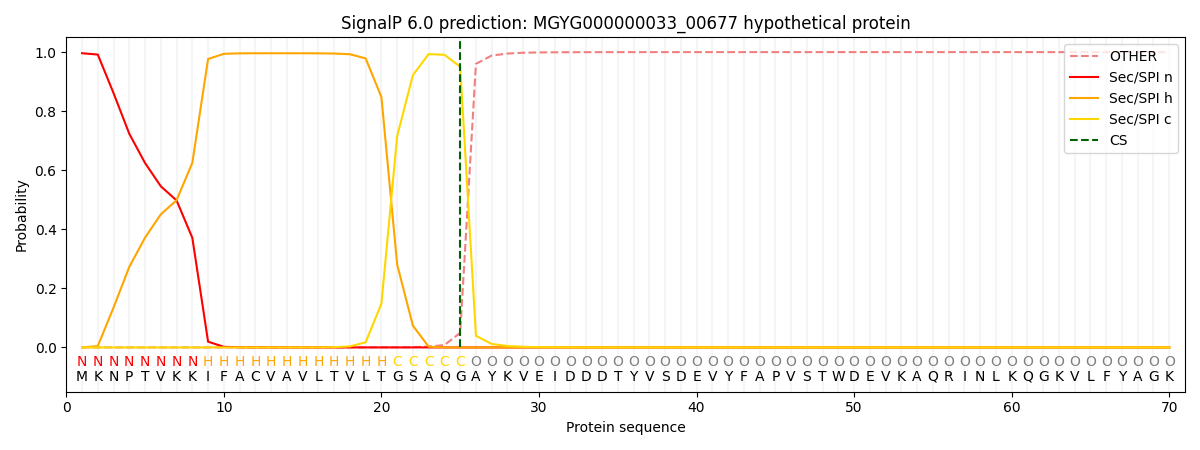
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000614 | 0.993657 | 0.004973 | 0.000252 | 0.000232 | 0.000228 |
