You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000033_01982
You are here: Home > Sequence: MGYG000000033_01982
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
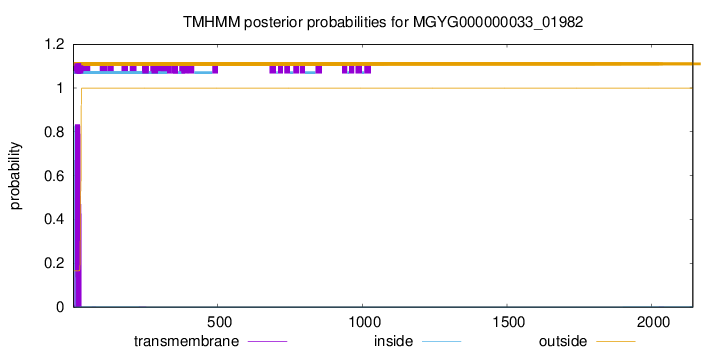
TMHMM annotations
Basic Information help
| Species | CAG-41 sp900066215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; CAG-41; CAG-41 sp900066215 | |||||||||||
| CAZyme ID | MGYG000000033_01982 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3703; End: 10134 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 53 | 360 | 6.2e-37 | 0.8763636363636363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 6.76e-22 | 63 | 348 | 8 | 270 | Cellulase (glycosyl hydrolase family 5). |
| pfam09479 | Flg_new | 2.58e-14 | 1976 | 2043 | 1 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| NF033189 | internalin_A | 9.35e-11 | 1837 | 2050 | 523 | 708 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| pfam09479 | Flg_new | 1.58e-10 | 1646 | 1713 | 1 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| NF033188 | internalin_H | 1.40e-08 | 1846 | 2048 | 282 | 484 | InlH/InlC2 family class 1 internalin. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin H (InlH), or internalin C2, two class 1 (LPXTG-type) internalins that are closely related, one apparently derived from the other through a recombination event. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT50938.1 | 5.24e-111 | 62 | 900 | 1 | 837 |
| QGT50946.1 | 2.08e-50 | 47 | 378 | 32 | 355 |
| ADR64665.1 | 1.61e-49 | 44 | 377 | 604 | 909 |
| AHF90980.1 | 1.58e-45 | 21 | 377 | 215 | 563 |
| QIF03557.1 | 1.57e-44 | 24 | 377 | 384 | 727 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HTY_A | 7.94e-17 | 43 | 381 | 47 | 358 | CrystalStructure of a metagenome-derived cellulase Cel5A [uncultured bacterium],4HU0_A Crystal Structure of a metagenome-derived cellulase Cel5A in complex with cellotetraose [uncultured bacterium] |
| 5A8N_A | 6.56e-16 | 47 | 378 | 51 | 360 | Crystalstructure of the native form of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40] |
| 5A8O_A | 1.57e-15 | 47 | 378 | 51 | 360 | Crystalstructure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with cellotetraose [Saccharophagus degradans 2-40],5A8P_A Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide B [Saccharophagus degradans 2-40],5A8Q_A Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A obtained by soaking [Saccharophagus degradans 2-40],5A94_A Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_B Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_C Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_D Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_E Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A94_F Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 [Saccharophagus degradans 2-40],5A95_A Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40],5A95_B Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40],5A95_C Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 [Saccharophagus degradans 2-40] |
| 5A8M_A | 9.07e-14 | 47 | 309 | 51 | 298 | Crystalstructure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40],5A8M_B Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40],5A8M_C Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans [Saccharophagus degradans 2-40] |
| 5WH8_A | 1.51e-10 | 47 | 377 | 14 | 307 | CellulaseCel5C_n [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DJ97 | 4.13e-20 | 1638 | 2064 | 423 | 817 | Putative Gly-rich membrane protein Bcell_0380 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0380 PE=4 SV=1 |
| P0DJ98 | 4.80e-17 | 1838 | 2088 | 41 | 262 | Putative membrane protein Bcell_0381 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0381 PE=4 SV=1 |
| P0DJM0 | 4.81e-13 | 1776 | 2050 | 461 | 709 | Internalin A OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=inlA PE=1 SV=1 |
| Q723K6 | 6.32e-13 | 1741 | 2050 | 410 | 709 | Internalin A OS=Listeria monocytogenes serotype 4b (strain F2365) OX=265669 GN=inlA PE=3 SV=1 |
| G2K3G6 | 1.09e-12 | 1776 | 2050 | 461 | 709 | Internalin A OS=Listeria monocytogenes serotype 1/2a (strain 10403S) OX=393133 GN=inlA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
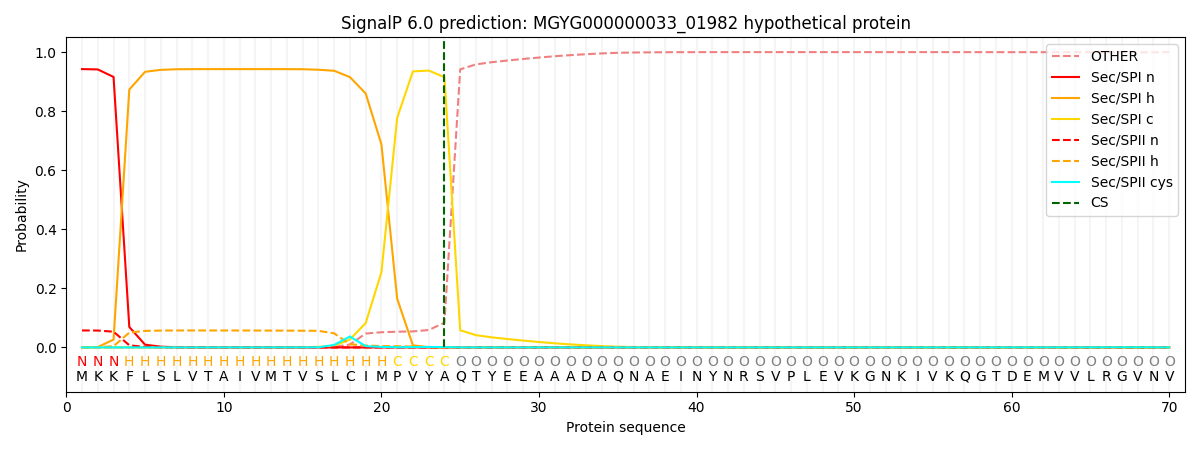
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000453 | 0.936709 | 0.062029 | 0.000300 | 0.000271 | 0.000223 |