You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000033_01989
You are here: Home > Sequence: MGYG000000033_01989
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-41 sp900066215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; CAG-41; CAG-41 sp900066215 | |||||||||||
| CAZyme ID | MGYG000000033_01989 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 26616; End: 29165 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 48 | 337 | 1.3e-33 | 0.8618181818181818 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 6.08e-22 | 49 | 328 | 1 | 267 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 0.002 | 117 | 260 | 116 | 270 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam08547 | CIA30 | 0.005 | 434 | 522 | 56 | 152 | Complex I intermediate-associated protein 30 (CIA30). This protein is associated with mitochondrial Complex I intermediate-associated protein 30 (CIA30) in human and mouse. The family is also present in Schizosaccharomyces pombe which does not contain the NADH dehydrogenase component of complex I, or many of the other essential subunits. This means it is possible that this family of protein may not be directly involved in oxidative phosphorylation. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGT50946.1 | 0.0 | 4 | 849 | 2 | 846 |
| QIF03557.1 | 1.70e-61 | 38 | 361 | 406 | 730 |
| AMV22357.1 | 3.10e-60 | 38 | 366 | 412 | 743 |
| QDU27947.1 | 1.39e-59 | 38 | 355 | 408 | 726 |
| ADR64665.1 | 1.03e-56 | 46 | 366 | 613 | 917 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HTY_A | 3.72e-13 | 41 | 361 | 52 | 357 | CrystalStructure of a metagenome-derived cellulase Cel5A [uncultured bacterium],4HU0_A Crystal Structure of a metagenome-derived cellulase Cel5A in complex with cellotetraose [uncultured bacterium] |
| 1A3H_A | 2.17e-12 | 40 | 328 | 8 | 260 | EndoglucanaseCel5a From Bacillus Agaradherans At 1.6a Resolution [Salipaludibacillus agaradhaerens],2A3H_A Cellobiose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 2.0 A Resolution [Salipaludibacillus agaradhaerens],3A3H_A Cellotriose Complex Of The Endoglucanase Cel5a From Bacillus Agaradherans At 1.6 A Resolution [Salipaludibacillus agaradhaerens] |
| 1H11_A | 2.25e-12 | 40 | 328 | 11 | 263 | 2-DEOXY-2-FLURO-B-D-CELLOTRIOSYL/ENZYMEINTERMEDIATE COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION [Salipaludibacillus agaradhaerens],1H2J_A ENDOGLUCANASE CEL5A IN COMPLEX WITH UNHYDROLYSED AND COVALENTLY LINKED 2,4-DINITROPHENYL-2-DEOXY-2-FLUORO-CELLOBIOSIDE AT 1.15 A RESOLUTION [Salipaludibacillus agaradhaerens],1HF6_A ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHAERENS IN THE ORTHORHOMBIC CRYSTAL FORM IN COMPLEX WITH CELLOTRIOSE [Salipaludibacillus agaradhaerens],1OCQ_A COMPLEX OF THE ENDOGLUCANASE CEL5A FROM BACILLUS AGARADHEARANS AT 1.08 ANGSTROM RESOLUTION with cellobio-derived isofagomine [Salipaludibacillus agaradhaerens],1W3K_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellobio Derived-tetrahydrooxazine [Salipaludibacillus agaradhaerens],1W3L_A Endoglucanase Cel5a From Bacillus Agaradhaerens In Complex With Cellotri Derived-Tetrahydrooxazine [Salipaludibacillus agaradhaerens],4A3H_A 2',4' Dinitrophenyl-2-Deoxy-2-Fluro-B-D-Cellobioside Complex Of The Endoglucanase Cel5a From Bacillus Agaradhaerens At 1.6 A Resolution [Salipaludibacillus agaradhaerens],5A3H_A 2-Deoxy-2-Fluro-B-D-CellobiosylENZYME INTERMEDIATE COMPLEX Of The Endoglucanase Cel5a From Bacillus Agaradhearans At 1.8 Angstroms Resolution [Salipaludibacillus agaradhaerens],6A3H_A 2-Deoxy-2-Fluro-B-D-CellotriosylENZYME INTERMEDIATE COMPLEX OF THE Endoglucanase Cel5a From Bacillus Agaradhearans At 1.6 Angstrom Resolution [Salipaludibacillus agaradhaerens],7A3H_A Native Endoglucanase Cel5a Catalytic Core Domain At 0.95 Angstroms Resolution [Salipaludibacillus agaradhaerens],8A3H_A Cellobiose-derived imidazole complex of the endoglucanase cel5A from Bacillus agaradhaerens at 0.97 A resolution [Salipaludibacillus agaradhaerens] |
| 1H5V_A | 2.27e-12 | 40 | 328 | 11 | 263 | Thiopentasaccharidecomplex of the endoglucanase Cel5A from Bacillus agaradharens at 1.1 A resolution in the tetragonal crystal form [Salipaludibacillus agaradhaerens] |
| 1E5J_A | 2.30e-12 | 40 | 328 | 11 | 263 | EndoglucanaseCel5a From Bacillus Agaradhaerens In The Tetragonal Crystal Form In Complex With Methyl-4ii-S-Alpha-Cellobiosyl-4ii-Thio Beta-Cellobioside [Salipaludibacillus agaradhaerens],1QHZ_A Native Tetragonal Structure Of The Endoglucanase Cel5a From Bacillus Agaradhaerens [Salipaludibacillus agaradhaerens],1QI0_A Endoglucanase Cel5a From Bacillus Agaradhaerens In The Tetragonal Crystal Form In Complex With Cellobiose [Salipaludibacillus agaradhaerens],1QI2_A Endoglucanase Cel5a From Bacillus Agaradhaerens In The Tetragonal Crystal Form In Complex With 2',4'-Dinitrophenyl 2-Deoxy-2-Fluoro-B- D-Cellotrioside [Salipaludibacillus agaradhaerens],2V38_A Family 5 endoglucanase Cel5A from Bacillus agaradhaerens in complex with cellobio-derived noeuromycin [Salipaludibacillus agaradhaerens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P07983 | 8.25e-14 | 7 | 328 | 9 | 292 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| O85465 | 1.13e-11 | 40 | 380 | 37 | 341 | Endoglucanase 5A OS=Salipaludibacillus agaradhaerens OX=76935 GN=cel5A PE=1 SV=1 |
| P10475 | 2.21e-11 | 7 | 288 | 9 | 259 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P06565 | 2.78e-11 | 40 | 368 | 37 | 336 | Endoglucanase B OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=celB PE=3 SV=1 |
| P23549 | 1.09e-09 | 7 | 288 | 9 | 259 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
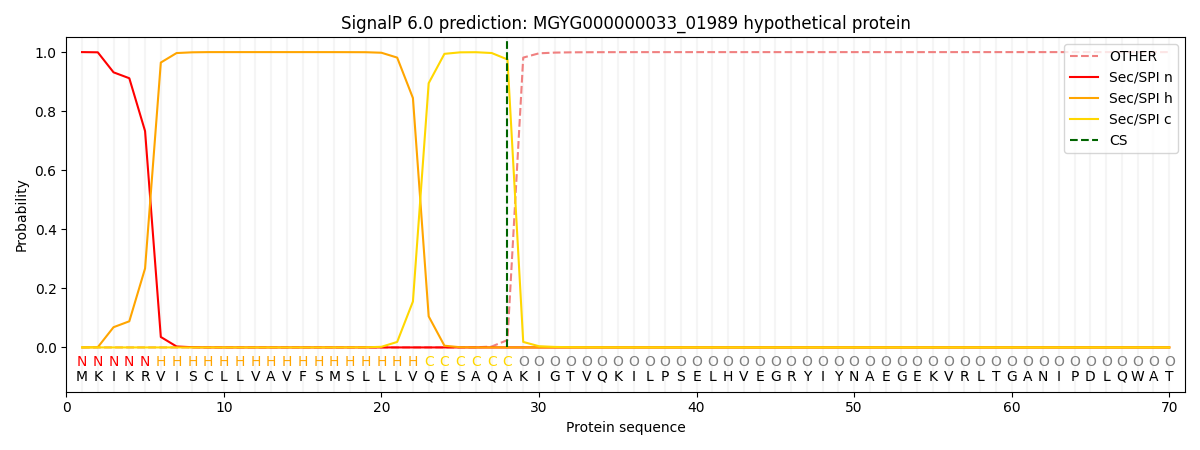
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000231 | 0.999087 | 0.000201 | 0.000165 | 0.000157 | 0.000142 |
