You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000033_02279
You are here: Home > Sequence: MGYG000000033_02279
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
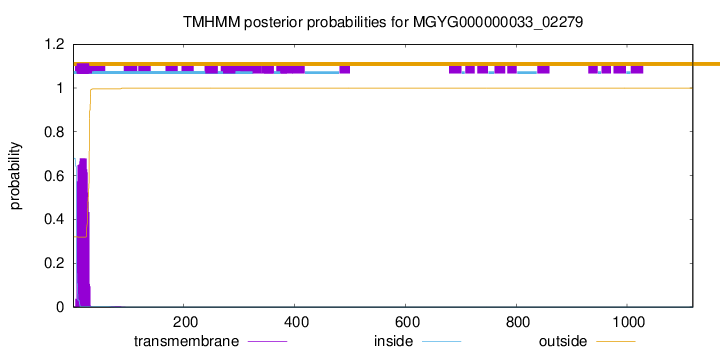
TMHMM annotations
Basic Information help
| Species | CAG-41 sp900066215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Monoglobales_A; UBA1381; CAG-41; CAG-41 sp900066215 | |||||||||||
| CAZyme ID | MGYG000000033_02279 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11860; End: 15219 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 36 | 572 | 3.7e-129 | 0.9905123339658444 |
| CBM32 | 656 | 775 | 1.2e-20 | 0.9032258064516129 |
| CBM32 | 890 | 1007 | 3.5e-20 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.92e-12 | 887 | 1006 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.03e-11 | 655 | 771 | 5 | 121 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam03442 | CBM_X2 | 1.03e-11 | 789 | 871 | 1 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
| cd00057 | FA58C | 0.001 | 890 | 969 | 16 | 95 | Substituted updates: Jan 31, 2002 |
| cd00057 | FA58C | 0.008 | 664 | 735 | 26 | 95 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGQ99051.1 | 5.05e-199 | 33 | 1010 | 36 | 915 |
| QGQ98753.1 | 1.19e-98 | 38 | 1011 | 36 | 1044 |
| QXE38906.1 | 1.24e-93 | 21 | 1010 | 22 | 1085 |
| QIQ07245.1 | 1.47e-87 | 27 | 1010 | 42 | 1100 |
| ADL27132.1 | 1.48e-87 | 28 | 582 | 22 | 592 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 1.73e-49 | 36 | 585 | 26 | 596 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
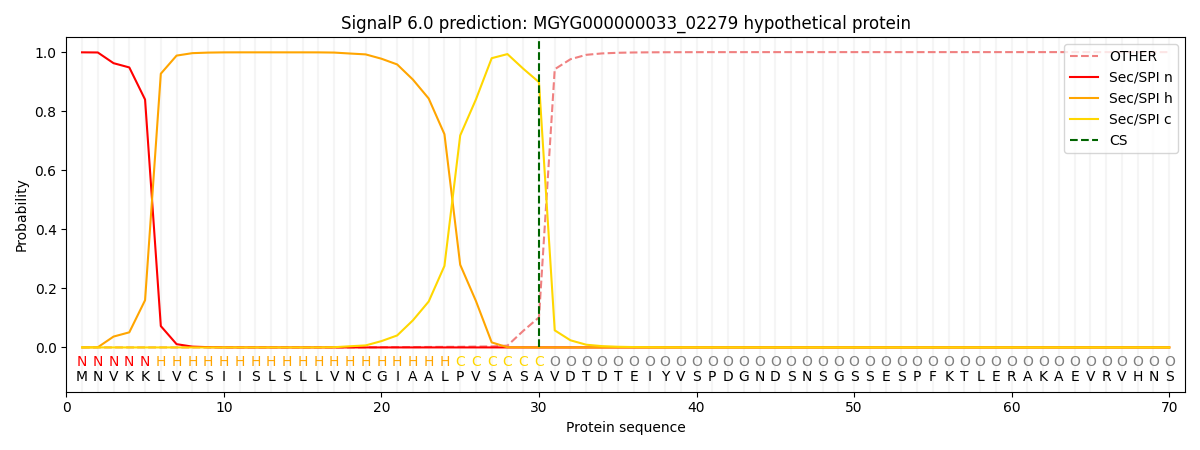
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000445 | 0.997674 | 0.001248 | 0.000224 | 0.000187 | 0.000176 |