You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000041_00099
You are here: Home > Sequence: MGYG000000041_00099
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Acetatifactor sp900066365 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; Acetatifactor sp900066365 | |||||||||||
| CAZyme ID | MGYG000000041_00099 | |||||||||||
| CAZy Family | CBM61 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 91576; End: 92139 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM61 | 39 | 184 | 1.4e-30 | 0.9858156028368794 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM03635.1 | 3.26e-136 | 1 | 187 | 1 | 187 |
| AEN97261.1 | 4.45e-104 | 1 | 187 | 1 | 185 |
| BBF43149.1 | 1.51e-30 | 8 | 186 | 9 | 180 |
| QDY72601.1 | 3.57e-30 | 39 | 186 | 668 | 812 |
| ARB18040.1 | 3.57e-30 | 39 | 187 | 668 | 813 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2XOM_A | 5.11e-10 | 39 | 187 | 9 | 151 | Atomicresolution structure of TmCBM61 in complex with beta-1,4- galactotriose [Thermotoga maritima],2XON_A Structure of TmCBM61 in complex with beta-1,4-galactotriose at 1.4 A resolution [Thermotoga maritima],2XON_L Structure of TmCBM61 in complex with beta-1,4-galactotriose at 1.4 A resolution [Thermotoga maritima] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
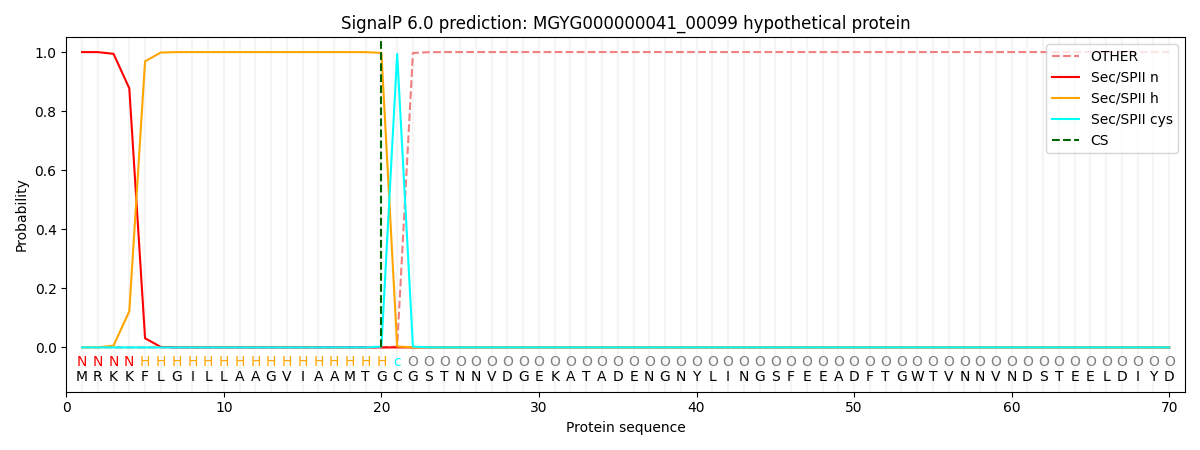
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000074 | 0.000000 | 0.000000 | 0.000000 |
