You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000042_00856
You are here: Home > Sequence: MGYG000000042_00856
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900066445 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900066445 | |||||||||||
| CAZyme ID | MGYG000000042_00856 | |||||||||||
| CAZy Family | GH18 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 243711; End: 246161 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH18 | 556 | 802 | 2.1e-38 | 0.8108108108108109 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00704 | Glyco_hydro_18 | 5.51e-33 | 578 | 794 | 53 | 307 | Glycosyl hydrolases family 18. |
| smart00636 | Glyco_18 | 2.06e-29 | 578 | 794 | 57 | 334 | Glyco_18 domain. |
| cd06548 | GH18_chitinase | 4.21e-28 | 552 | 794 | 41 | 322 | The GH18 (glycosyl hydrolases, family 18) type II chitinases hydrolyze chitin, an abundant polymer of N-acetylglucosamine and have been identified in bacteria, fungi, insects, plants, viruses, and protozoan parasites. The structure of this domain is an eight-stranded alpha/beta barrel with a pronounced active-site cleft at the C-terminal end of the beta-barrel. |
| cd06545 | GH18_3CO4_chitinase | 5.67e-21 | 565 | 807 | 31 | 253 | The Bacteroides thetaiotaomicron protein represented by pdb structure 3CO4 is an uncharacterized bacterial member of the family 18 glycosyl hydrolases with homologs found in Flavobacterium, Stigmatella, and Pseudomonas. |
| COG3325 | ChiA | 5.62e-19 | 578 | 794 | 117 | 423 | Chitinase, GH18 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT71950.1 | 3.36e-134 | 319 | 811 | 61 | 545 |
| QQA10039.1 | 3.36e-134 | 319 | 811 | 61 | 545 |
| QMW88601.1 | 3.36e-134 | 319 | 811 | 61 | 545 |
| AAO76739.1 | 3.36e-134 | 319 | 811 | 61 | 545 |
| ALJ40996.1 | 3.36e-134 | 319 | 811 | 61 | 545 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1ITX_A | 3.03e-14 | 555 | 794 | 95 | 406 | ChainA, Glycosyl Hydrolase [Niallia circulans] |
| 6TSB_AAA | 6.65e-12 | 578 | 794 | 78 | 337 | ChainAAA, Peroxiredoxin [Clostridioides difficile 630] |
| 6T9M_AAA | 7.48e-12 | 578 | 794 | 97 | 356 | ChainAAA, Peroxiredoxin [Clostridioides difficile 630] |
| 3AQU_A | 4.60e-11 | 586 | 794 | 64 | 330 | Crystalstructure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana],3AQU_B Crystal structure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana],3AQU_C Crystal structure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana],3AQU_D Crystal structure of a class V chitinase from Arabidopsis thaliana [Arabidopsis thaliana] |
| 6EPB_A | 8.61e-11 | 584 | 794 | 117 | 383 | Structureof Chitinase 42 from Trichoderma harzianum [Trichoderma harzianum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E9ERT9 | 2.32e-14 | 505 | 794 | 24 | 384 | Endochitinase 1 OS=Metarhizium robertsii (strain ARSEF 23 / ATCC MYA-3075) OX=655844 GN=chit1 PE=3 SV=1 |
| P20533 | 3.55e-13 | 555 | 794 | 127 | 438 | Chitinase A1 OS=Niallia circulans OX=1397 GN=chiA1 PE=1 SV=1 |
| O14456 | 2.19e-12 | 505 | 794 | 24 | 383 | Endochitinase 1 OS=Metarhizium anisopliae OX=5530 GN=chit1 PE=1 SV=1 |
| P48827 | 2.02e-10 | 584 | 794 | 117 | 383 | Endochitinase 42 OS=Trichoderma harzianum OX=5544 GN=chit42 PE=1 SV=1 |
| O81862 | 2.89e-10 | 586 | 794 | 87 | 353 | Class V chitinase OS=Arabidopsis thaliana OX=3702 GN=ChiC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
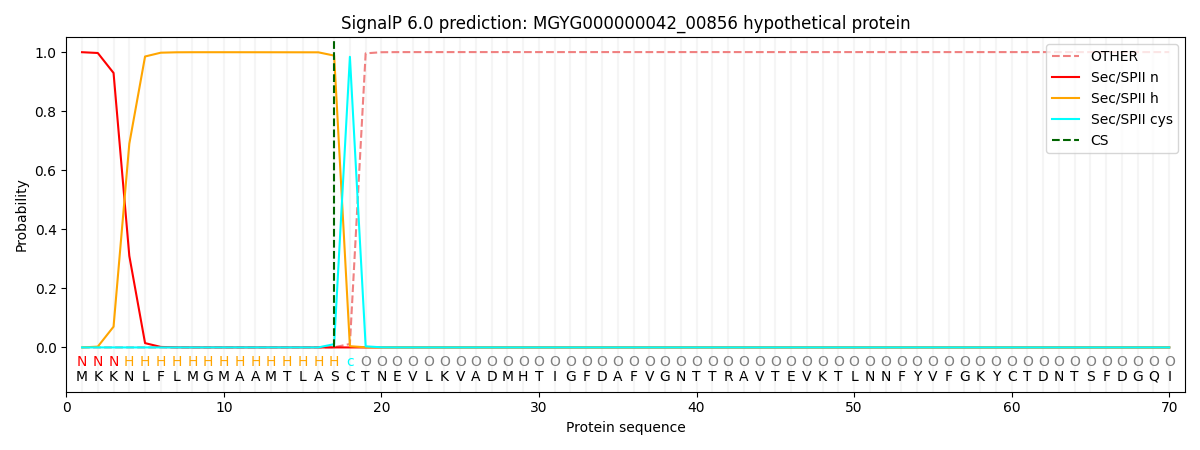
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000389 | 0.999656 | 0.000000 | 0.000000 | 0.000000 |
