You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000043_03309
You are here: Home > Sequence: MGYG000000043_03309
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Phocaeicola sp900066455 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Phocaeicola; Phocaeicola sp900066455 | |||||||||||
| CAZyme ID | MGYG000000043_03309 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 423; End: 2930 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 43 | 754 | 1.3e-61 | 0.9778393351800554 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1554 | ATH1 | 8.68e-05 | 206 | 481 | 190 | 484 | Trehalose and maltose hydrolase (possible phosphorylase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 3.79e-33 | 147 | 796 | 465 | 1081 |
| AZU63742.1 | 3.09e-30 | 55 | 822 | 60 | 832 |
| CBL25699.1 | 8.20e-29 | 321 | 828 | 408 | 915 |
| SMD45424.1 | 8.53e-29 | 57 | 758 | 50 | 736 |
| QRO25182.1 | 4.23e-28 | 42 | 777 | 29 | 745 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RDY_A | 1.13e-07 | 441 | 801 | 458 | 794 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 4UFC_A | 6.84e-06 | 320 | 796 | 332 | 804 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
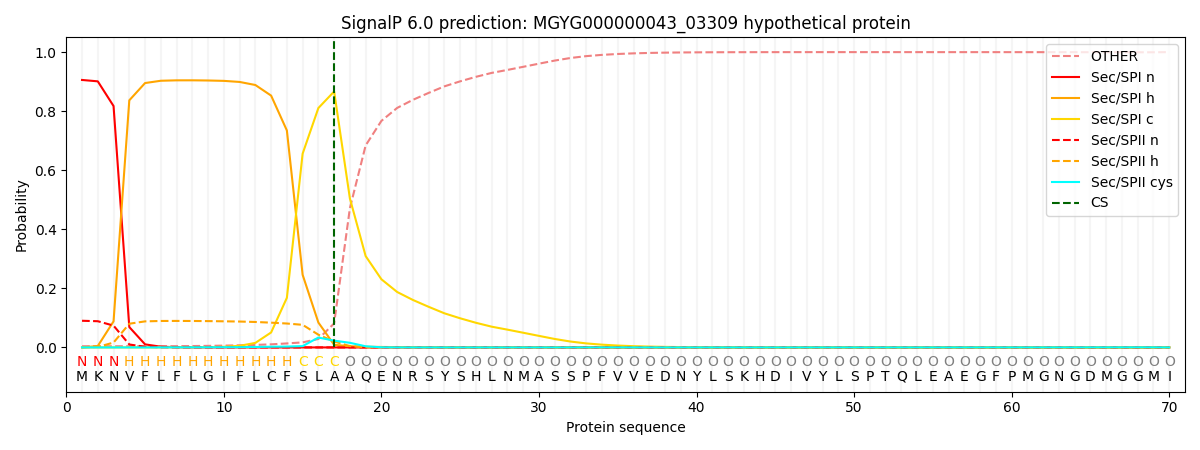
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.005627 | 0.900250 | 0.093336 | 0.000270 | 0.000246 | 0.000254 |
