You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000048_00926
You are here: Home > Sequence: MGYG000000048_00926
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Clostridium_P ventriculi | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Clostridiales; Clostridiaceae; Clostridium_P; Clostridium_P ventriculi | |||||||||||
| CAZyme ID | MGYG000000048_00926 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16232; End: 17476 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08239 | SH3_3 | 5.56e-12 | 276 | 327 | 1 | 54 | Bacterial SH3 domain. |
| COG3103 | YgiM | 2.13e-10 | 276 | 414 | 32 | 152 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| pfam08239 | SH3_3 | 3.99e-08 | 357 | 412 | 1 | 54 | Bacterial SH3 domain. |
| smart00287 | SH3b | 4.75e-06 | 268 | 329 | 2 | 63 | Bacterial SH3 domain homologues. |
| PRK13914 | PRK13914 | 1.04e-05 | 276 | 327 | 87 | 140 | invasion associated endopeptidase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QPJ84712.1 | 1.17e-286 | 1 | 414 | 1 | 414 |
| QLY80841.1 | 2.09e-86 | 51 | 412 | 379 | 759 |
| QAA30551.1 | 1.25e-63 | 68 | 412 | 46 | 423 |
| QOR36547.1 | 8.80e-33 | 69 | 243 | 157 | 334 |
| ASN72389.1 | 2.46e-32 | 99 | 246 | 290 | 448 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2KRS_A | 4.97e-15 | 270 | 333 | 3 | 66 | SolutionNMR structure of SH3 domain from CPF_0587 (fragment 415-479) from Clostridium perfringens. Northeast Structural Genomics Consortium (NESG) Target CpR74A. [Clostridium perfringens] |
| 2KT8_A | 3.11e-13 | 268 | 326 | 3 | 61 | SolutionNMR structure of the CPE1231(468-535) protein from Clostridium perfringens, Northeast Structural Genomics Consortium Target CpR82B [Clostridium perfringens] |
| 2KYB_A | 1.85e-12 | 268 | 326 | 2 | 60 | Solutionstructure of CpR82G from Clostridium perfringens. North East Structural Genomics Consortium Target CpR82g [Clostridium perfringens ATCC 13124] |
| 2KQ8_A | 2.10e-08 | 277 | 329 | 11 | 63 | ChainA, Cell wall hydrolase [[Bacillus thuringiensis] serovar konkukian] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 1.04e-20 | 57 | 213 | 26 | 179 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
| O32041 | 1.04e-12 | 271 | 411 | 105 | 239 | Putative N-acetylmuramoyl-L-alanine amidase YrvJ OS=Bacillus subtilis (strain 168) OX=224308 GN=yrvJ PE=3 SV=1 |
| P08696 | 4.63e-07 | 200 | 333 | 512 | 639 | Bacteriocin BCN5 OS=Clostridium perfringens OX=1502 GN=bcn PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
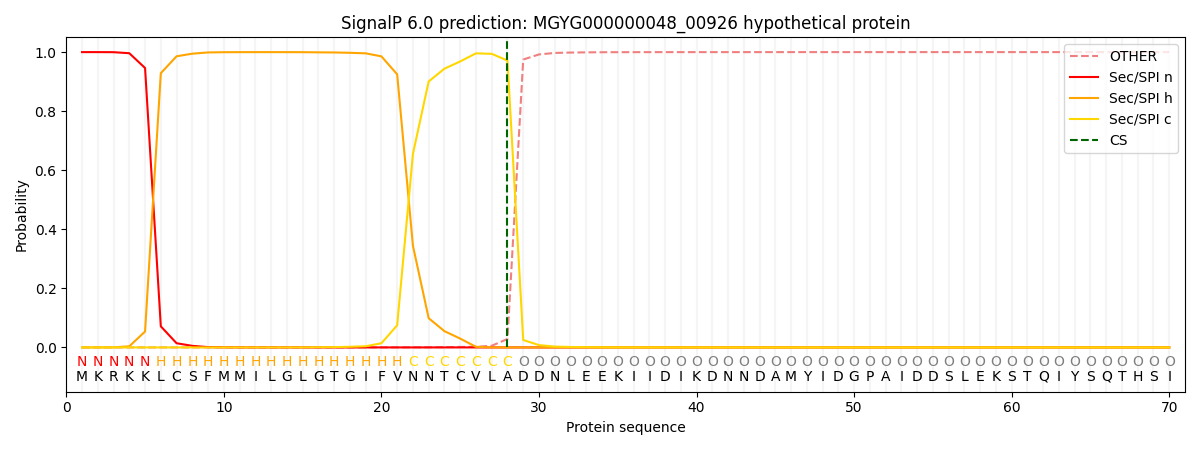
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000464 | 0.998735 | 0.000291 | 0.000180 | 0.000157 | 0.000147 |
