You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000049_00106
You are here: Home > Sequence: MGYG000000049_00106
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | GCA-900066495 sp900066495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; GCA-900066495; GCA-900066495 sp900066495 | |||||||||||
| CAZyme ID | MGYG000000049_00106 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 118545; End: 124877 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 1763 | 1903 | 4.2e-43 | 0.9925373134328358 |
| CBM51 | 1536 | 1676 | 3.1e-42 | 0.9925373134328358 |
| CBM51 | 1316 | 1450 | 9.1e-33 | 0.9850746268656716 |
| CBM32 | 74 | 202 | 7.3e-16 | 0.9435483870967742 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 1.54e-50 | 1761 | 1904 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 1.15e-46 | 1534 | 1676 | 1 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 1.31e-42 | 1762 | 1904 | 4 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| smart00776 | NPCBM | 1.90e-42 | 1533 | 1676 | 2 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| pfam08305 | NPCBM | 6.92e-38 | 1316 | 1451 | 4 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SQG38452.1 | 0.0 | 1 | 1990 | 1 | 1984 |
| AXH52096.1 | 0.0 | 1 | 1990 | 1 | 1984 |
| AMN35289.1 | 0.0 | 1 | 1990 | 1 | 1984 |
| ABG84962.1 | 0.0 | 1 | 1990 | 1 | 1970 |
| AMN32462.1 | 0.0 | 1 | 1990 | 1 | 1970 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JFS_A | 1.48e-172 | 272 | 1233 | 27 | 980 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JS4_A | 1.90e-130 | 717 | 1676 | 23 | 975 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JND_A | 2.67e-118 | 18 | 446 | 22 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124],7JNF_A Chain A, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JNB_A | 1.09e-116 | 18 | 446 | 22 | 449 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 5KDJ_A | 1.28e-75 | 652 | 1312 | 24 | 672 | ZmpBmetallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124],5KDJ_B ZmpB metallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6RUF5 | 7.42e-06 | 1764 | 1903 | 43 | 203 | Blood-group-substance endo-1,4-beta-galactosidase OS=Clostridium perfringens OX=1502 GN=eabC PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
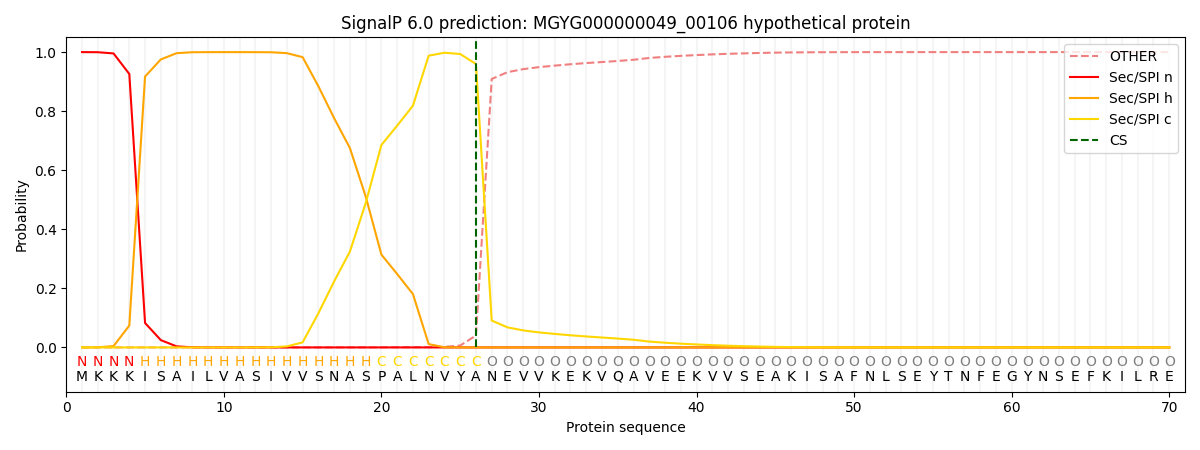
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000384 | 0.998891 | 0.000182 | 0.000192 | 0.000166 | 0.000146 |
