You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000049_03280
You are here: Home > Sequence: MGYG000000049_03280
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | GCA-900066495 sp900066495 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; GCA-900066495; GCA-900066495 sp900066495 | |||||||||||
| CAZyme ID | MGYG000000049_03280 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14411; End: 16855 Strand: - | |||||||||||
Full Sequence Download help
| MGFVVSNGGS TTPAVANFND ITIKNESGII YTSTDIQTPE VPEVPEIPET PELPNYGVGT | 60 |
| LPDGFNNSSI GNDSENVYVN FDKDTKSFTV NGSGTYIGKD TSSTDNYQFV NYKVEGNATI | 120 |
| VARLVDFDMT NAKYGQAGVF IRNNNFTNDA DYFGVYVEPS KNQYRYAYRD NSSAKSGAAQ | 180 |
| IGELNADSKN QYIKIERNKT QFKAYISKDP TFPTDKTIVK TQTVVNESNT WYVGFVVSNG | 240 |
| GSETPAVDTF DNVRIENEDK LYYDSTIEKM PVDTVENVKA EAKDFKVTLS WDEVKDATSY | 300 |
| VIKRSTEKNG EYKEIAEVDS SKTTYEDLEV VNFQTYYYKI MAKNENGNAY DSKEVVAVPN | 360 |
| NSNPDNIQYE DNAANFNMIE EPNDTVSNPV ITLKGSVDKD GKITIKQNDK IKVNAIEKSK | 420 |
| DEVFTKNLTL DLGRNIIEIY HTTEDGKTTV KTYNIVYLNN GNYDIIVDST YNGVEGREVD | 480 |
| GIKTYSTITE AVDSISTKNT ERVNIFVKNG VYKEKTIVQS PYISIIGEDS QKTIWTYDVS | 540 |
| NGTINPETGE KYGTSKSASV TIKSKAVGFT AENITIENAY EEKQVVEGDQ AVALNNQADQ | 600 |
| SIFINCRFIG NQDTLLADAS SSLPARQYYY KCYIEGDVDF IFVRAQAVFN DCDIASVNRN | 660 |
| LSPKNGYITA ADTWDKDAYG YVIMNSRLIG LDDIADNSVS LGRPWRPSSA TDTVTPAVAY | 720 |
| INCYMGSHIA TNGWDDMGAN SLASTARFFE YGSYGPGAKL SDTKPLIQNS EIANYSLENV | 780 |
| FSTSSASVNG NDAYTTNWNP TEKSDSTNIN SAYG | 814 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 2.80e-53 | 469 | 782 | 79 | 403 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 2.24e-48 | 480 | 761 | 7 | 269 | Pectinesterase. |
| PLN02432 | PLN02432 | 5.50e-46 | 485 | 761 | 23 | 263 | putative pectinesterase |
| PLN02682 | PLN02682 | 8.97e-41 | 485 | 759 | 82 | 336 | pectinesterase family protein |
| PLN02671 | PLN02671 | 3.64e-40 | 452 | 758 | 48 | 325 | pectinesterase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CED92996.1 | 0.0 | 2 | 813 | 211 | 1037 |
| QJS18268.1 | 1.21e-295 | 60 | 813 | 118 | 875 |
| BCS80168.1 | 2.41e-93 | 87 | 781 | 559 | 1241 |
| ADQ45070.1 | 1.08e-86 | 83 | 780 | 557 | 1240 |
| ADQ05621.1 | 1.08e-81 | 98 | 780 | 575 | 1241 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2NSP_A | 9.23e-37 | 469 | 780 | 2 | 333 | ChainA, Pectinesterase A [Dickeya dadantii 3937],2NSP_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NST_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NST_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT6_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT6_B Chain B, Pectinesterase A [Dickeya dadantii 3937],2NT9_A Chain A, Pectinesterase A [Dickeya dadantii 3937],2NT9_B Chain B, Pectinesterase A [Dickeya dadantii 3937] |
| 1QJV_A | 4.70e-33 | 469 | 780 | 2 | 333 | ChainA, PECTIN METHYLESTERASE [Dickeya chrysanthemi],1QJV_B Chain B, PECTIN METHYLESTERASE [Dickeya chrysanthemi] |
| 2NTB_A | 4.70e-33 | 469 | 780 | 2 | 333 | Crystalstructure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTB_B Crystal structure of pectin methylesterase in complex with hexasaccharide V [Dickeya dadantii 3937],2NTP_A Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTP_B Crystal structure of pectin methylesterase in complex with hexasaccharide VI [Dickeya dadantii 3937],2NTQ_A Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937],2NTQ_B Crystal structure of pectin methylesterase in complex with hexasaccharide VII [Dickeya dadantii 3937] |
| 5C1E_A | 1.07e-28 | 484 | 762 | 18 | 270 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Penultimately Deglycosylated Form (N-acetylglucosamine Stub at Asn84) [Aspergillus niger ATCC 1015] |
| 5C1C_A | 1.45e-28 | 484 | 762 | 18 | 270 | CrystalStructure of the Pectin Methylesterase from Aspergillus niger in Deglycosylated Form [Aspergillus niger ATCC 1015] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FM79 | 1.63e-32 | 487 | 765 | 94 | 353 | Pectinesterase QRT1 OS=Arabidopsis thaliana OX=3702 GN=QRT1 PE=1 SV=1 |
| P0C1A9 | 4.17e-32 | 469 | 780 | 26 | 357 | Pectinesterase A OS=Dickeya dadantii (strain 3937) OX=198628 GN=pemA PE=1 SV=1 |
| P0C1A8 | 5.65e-32 | 469 | 780 | 26 | 357 | Pectinesterase A OS=Dickeya chrysanthemi OX=556 GN=pemA PE=1 SV=1 |
| O23038 | 1.25e-28 | 481 | 768 | 97 | 366 | Probable pectinesterase 8 OS=Arabidopsis thaliana OX=3702 GN=PME8 PE=2 SV=2 |
| Q8L7Q7 | 1.66e-28 | 485 | 765 | 301 | 573 | Probable pectinesterase/pectinesterase inhibitor 64 OS=Arabidopsis thaliana OX=3702 GN=PME64 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
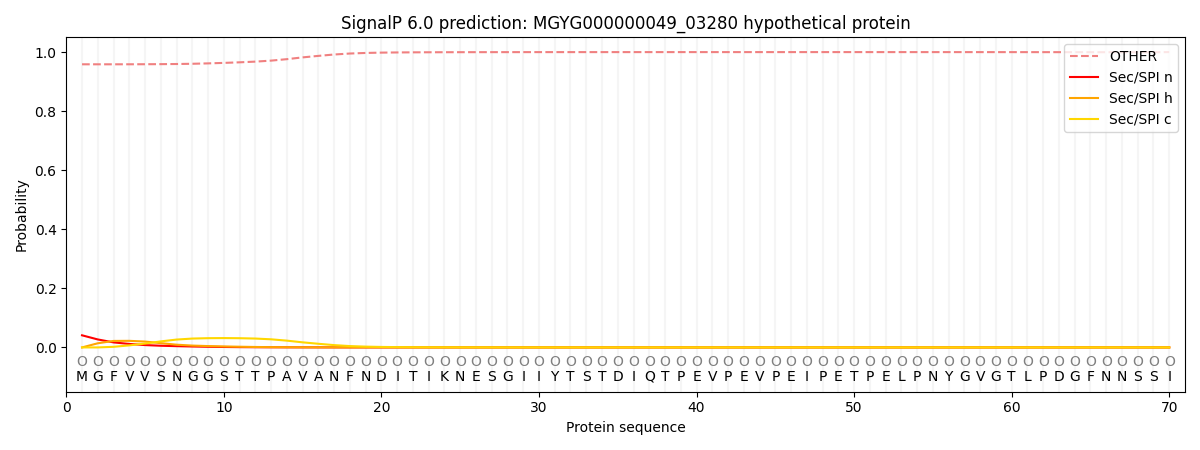
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.961675 | 0.037737 | 0.000319 | 0.000118 | 0.000048 | 0.000122 |
