You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000051_01381
You are here: Home > Sequence: MGYG000000051_01381
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
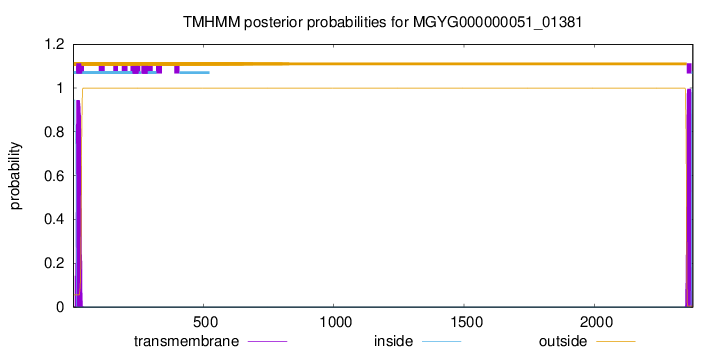
TMHMM annotations
Basic Information help
| Species | Anaerosacchariphilus sp900066385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerosacchariphilus; Anaerosacchariphilus sp900066385 | |||||||||||
| CAZyme ID | MGYG000000051_01381 | |||||||||||
| CAZy Family | GH31 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 116998; End: 124134 Strand: - | |||||||||||
Full Sequence Download help
| MIKKKKMSSK FLSIVLAAAM VAGIAPQFPA STVEAAFNGP LSSVKSARAN DNVVEVSFNN | 60 |
| GDVKGKITFL EDGIFRYNVD PSGKFSEYAT VNNAADTAKI PQYPDSSDKY SHPKAKVSDE | 120 |
| NGNIVIAAGK TKITFDKDTA KMTVAYNDNV VMQESAPLSL GTKTTQSLVK NSGENFYGGG | 180 |
| TQNGRFVHTG ETIHIANESN WVDGGVASPN PFYYSSNGYG VLRNTFLAGS YDFGNNSADT | 240 |
| VATAHNENEY DAYIFLSDGA NAAERVQDLL GEYYHVTGAP VLLPEYAFYE GHLNCYNRDS | 300 |
| WSDNSGSKKW EIKGSASATT ADEVPIASSY YESGMSTGYV LKDGMSSETL NGTKPTTALD | 360 |
| KYPSVNTDSK YSARAVIDQY DKYDMPIGYF LPNDGYGCGY GQNGYYQTGG VNADGSSSAE | 420 |
| RTAAVDANVN NLKEFTKYAQ SKGLQTGLWT QSNLSPDSNS NTSWHLLRDF EKEVKVGGIT | 480 |
| TLKTDVAWVG YGYNFALNGT KTAYDIVTTG VSKRPNIITL DGWASTQRFG AIWTGDQYGG | 540 |
| NWEYIRFHVP TYIGQSLSGN PNIGSDMDGI FGGDKIVSTR DTQWKVFTPL MLNMDGWGSY | 600 |
| AKTPQTFGDP YTGINRMYLK LKAQLMPYIY TSAAAAANLD TGNGDTGLPM IRAMFLEYPG | 660 |
| DAYASTKDMQ YQYMFGSNVL VAPIYQNTSS DTQGNDVRNG IYLPDADQIW IDYFTGEQYY | 720 |
| GGQTLNNFDA PIWKLPIFVK NGSIIPMWEE NNSPSKINKA NRSVEFWPAG STEYTMYEDD | 780 |
| GITAENETTE VDGYGTVADV SYGEHVSQKF TSKVEGNVAT LTAEKASGSY AGYDKNKNTT | 840 |
| FVVNVSKEPT AITAQNGSTE LTKVAMKSKD EVLNADVAAG TFAYYYDESP AIEYYGVEAE | 900 |
| DEFAAMMEAQ TSSPKLYVKF ANTDSQANAQ TLVLEGFENN GDMSKNQLNP ALAAPAKLTD | 960 |
| DVTRKTPTSN TLTWDAVDGA TSYELLVDGI INTAGDATEF AHTDLKYNST HTYQVRSRNK | 1020 |
| DGYSEWSEEV TATTSLDPWR NVPTPEKITW TGGDSWGKLA NLTDHDISDA AGMFHSTGDV | 1080 |
| VTDAIPMIFD YGQAYALDKL EYYARHDNYG NGAVQQMNVE TSLDGVNWTT QWIGSEHSDW | 1140 |
| TYDAKASIAD NMKEIPLTGA ARYVKIVITK SKGNFFSGDE LAIYKKDGTN GFAVGSTNNL | 1200 |
| GTVADGDYTN MKNYLGFSKK DGETFVKQIE ERFGDINGNG YYDVWDYAFT MFNLDGGTQK | 1260 |
| TGKVSGNIVL LPSATDVKAG ETFTVDVYAD NVKNLNAFGE VLDYNPAVLQ YVDTTADFDI | 1320 |
| SQMENLTVNK VYEDGTAYVN LAFANRGNQA TYSGTGVIAT ITMKAKNDFT LDAKAMDLSQ | 1380 |
| VMLIGPEFDL INCESSSTPE LPDIVTGGEK TYTYGEDFDM TMTNNILKED DGKNVEKFIE | 1440 |
| SNSYDGLFNG SQGREFELKW YPNDWTTTVK GLPEHVKVPV TFHIDVKQDA ADEVVEDTTE | 1500 |
| DVADEAEDVE VQTAEEEVAV QADNTRKVTS VTLQNANKAN GYVTSASAEF TYSDGSTSSD | 1560 |
| SKSLDWATAA DYSEFTFNAE ADKTVTKVDV TVNSVANAKG ASVENMLTLS TLTVATADGK | 1620 |
| LSYGSDFDMT MTNAVLTSDD GDNVKKMIQQ GSFSGLFGGQ GRNFEFLWDI PSNYVDKEVK | 1680 |
| YVEMPVTLHM NMKKADTVST VSVYNAPSYG NGYMTDATAT FTYEDGTTAD VNLKDQGAKF | 1740 |
| EFANPNKDAK VTKVSVTANQ ASGTNMLTLS ELEVKAKVDM VPVTKITPAE DNVKELYAGS | 1800 |
| MADINATIEP ENATNPYFVA ASSNENVVRI ITLADENGLP IYKAYAVGEG KAKITLTARE | 1860 |
| SMQDTATQAE GEDPAAPITA SYEITVLAGA DKTELVELIT EYSKTNPKFY TEESFKALTD | 1920 |
| AINAANAVKD NAEATKDDVK KAIAAMKKAA DALEESELTA EAITDKSGMT ADAPYSEENV | 1980 |
| PSALVDGDET NYWESPYYGS NAGLPKEITL ALGDAYDLEK VEIVKNTGNG KVTDYEVLVS | 2040 |
| TDGENFTSVG RKTIDENEMR SGIRMEQANI THVKVIIHGA KTGGGDVAYA RLSEINFYGK | 2100 |
| KVAEPADKAV LTEAVDKYKD LKQENYTDET WAAFAEALKT AQNVLADENA TQETVDAAVK | 2160 |
| TLNDAYKGLV KKELPNPDQP VVKDALKAAV DKNADKKQGD YTADSWAKFS AALAHAQAVL | 2220 |
| NDQNATADQV SAALKGLEEA AAALTTGQSP AAKDELQAVY NGNKDREKKD YTADSWKAFS | 2280 |
| TALSNAEKIL KNPNATSQEI SDALNALKTA VADLKTVAEK KDEDNKKQNS SGSSGSSKSN | 2340 |
| TTSTTSAQTG DTAQPLLYVV LIAAAAAVLA LIRRKRMK | 2378 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH31 | 264 | 745 | 4.7e-57 | 0.9836065573770492 |
| CBM32 | 1974 | 2083 | 7.2e-16 | 0.8145161290322581 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd06596 | GH31_CPE1046 | 1.44e-152 | 276 | 716 | 1 | 334 | Clostridium CPE1046-like. CPE1046 is an uncharacterized Clostridium perfringens protein with a glycosyl hydrolase family 31 (GH31) domain. The domain architecture of CPE1046 and its orthologs includes a C-terminal fibronectin type 3 (FN3) domain and a coagulation factor 5/8 type C domain in addition to the GH31 domain. Enzymes of the GH31 family possess a wide range of different hydrolytic activities including alpha-glucosidase (glucoamylase and sucrase-isomaltase), alpha-xylosidase, 6-alpha-glucosyltransferase, 3-alpha-isomaltosyltransferase and alpha-1,4-glucan lyase. All GH31 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. |
| COG1501 | YicI | 5.79e-60 | 40 | 790 | 19 | 716 | Alpha-glucosidase, glycosyl hydrolase family GH31 [Carbohydrate transport and metabolism]. |
| pfam01055 | Glyco_hydro_31 | 3.16e-53 | 491 | 745 | 210 | 442 | Glycosyl hydrolases family 31. Glycosyl hydrolases are key enzymes of carbohydrate metabolism. Family 31 comprises of enzymes that are, or similar to, alpha- galactosidases. |
| cd06603 | GH31_GANC_GANAB_alpha | 1.95e-40 | 513 | 783 | 212 | 467 | neutral alpha-glucosidase C, neutral alpha-glucosidase AB. This subgroup includes the closely related glycosyl hydrolase family 31 (GH31) isozymes, neutral alpha-glucosidase C (GANC) and the alpha subunit of heterodimeric neutral alpha-glucosidase AB (GANAB). Initially distinguished on the basis of differences in electrophoretic mobility in starch gel, GANC and GANAB have been shown to have other differences, including those of substrate specificity. GANC and GANAB are key enzymes in glycogen metabolism that hydrolyze terminal, non-reducing 1,4-linked alpha-D-glucose residues from glycogen in the endoplasmic reticulum. The GANC/GANAB family includes the alpha-glucosidase II (ModA) from Dictyostelium discoideum as well as the alpha-glucosidase II (GLS2, or ROT2 - Reversal of TOR2 lethality protein 2) from Saccharomyces cerevisiae. |
| cd08759 | Type_III_cohesin_like | 1.13e-34 | 1266 | 1435 | 1 | 167 | Cohesin domain, interaction partner of dockerin. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. Two specific calcium-dependent interactions between cohesin and dockerin appear to be essential for cellulosome assembly, type I and type II. This subfamily represents type III cohesins and closely related domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBK61154.1 | 0.0 | 36 | 2308 | 38 | 2130 |
| BCT46261.1 | 0.0 | 38 | 2324 | 40 | 2125 |
| QUO30799.1 | 0.0 | 52 | 1390 | 38 | 1368 |
| QNM10857.1 | 0.0 | 39 | 2244 | 45 | 2011 |
| QWT17625.1 | 0.0 | 3 | 2250 | 10 | 2105 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6M76_A | 2.00e-247 | 39 | 1040 | 38 | 963 | GH31alpha-N-acetylgalactosaminidase from Enterococcus faecalis [Enterococcus faecalis ATCC 10100],6M77_A GH31 alpha-N-acetylgalactosaminidase from Enterococcus faecalis in complex with N-acetylgalactosamine [Enterococcus faecalis ATCC 10100] |
| 7F7R_A | 1.06e-246 | 39 | 1040 | 38 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 7F7Q_A | 2.87e-246 | 39 | 1040 | 38 | 963 | ChainA, GH31 alpha-N-acetylgalactosaminidase [Enterococcus faecalis ATCC 10100] |
| 5F7U_A | 2.00e-25 | 117 | 907 | 180 | 932 | Cycloalternan-formingenzyme from Listeria monocytogenes in complex with pentasaccharide substrate [Listeria monocytogenes EGD-e] |
| 6JR8_A | 3.37e-25 | 512 | 787 | 463 | 733 | Flavobacteriumjohnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_B Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_C Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101],6JR8_D Flavobacterium johnsoniae GH31 dextranase, FjDex31A, mutant D412A complexed with isomaltotriose [Flavobacterium johnsoniae UW101] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9F234 | 4.82e-33 | 513 | 816 | 461 | 743 | Alpha-glucosidase 2 OS=Bacillus thermoamyloliquefaciens OX=1425 PE=3 SV=1 |
| Q9P999 | 1.69e-26 | 489 | 787 | 376 | 664 | Alpha-xylosidase OS=Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) OX=273057 GN=xylS PE=1 SV=1 |
| Q9FN05 | 7.29e-23 | 447 | 746 | 493 | 780 | Probable glucan 1,3-alpha-glucosidase OS=Arabidopsis thaliana OX=3702 GN=PSL5 PE=1 SV=1 |
| P79403 | 3.38e-21 | 506 | 788 | 588 | 856 | Neutral alpha-glucosidase AB OS=Sus scrofa OX=9823 GN=GANAB PE=1 SV=1 |
| Q4R4N7 | 4.44e-21 | 509 | 788 | 591 | 856 | Neutral alpha-glucosidase AB OS=Macaca fascicularis OX=9541 GN=GANAB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
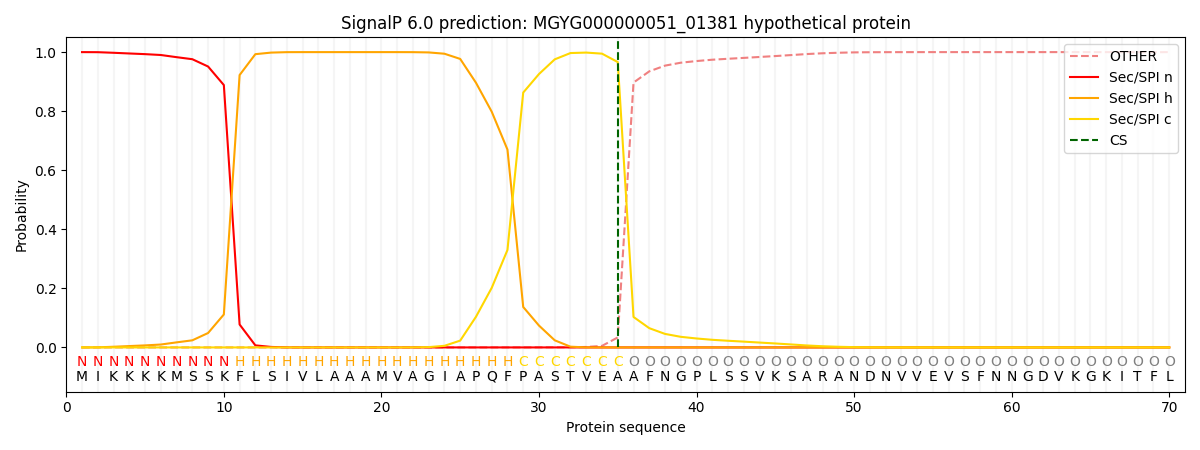
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000488 | 0.998561 | 0.000223 | 0.000272 | 0.000227 | 0.000191 |