You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000052_02673
You are here: Home > Sequence: MGYG000000052_02673
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Anaerostipes sp000508985 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Anaerostipes; Anaerostipes sp000508985 | |||||||||||
| CAZyme ID | MGYG000000052_02673 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 40659; End: 42044 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 9.99e-28 | 181 | 461 | 21 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 4.14e-05 | 264 | 294 | 7 | 37 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCP36275.1 | 9.45e-291 | 1 | 461 | 4 | 468 |
| BCD36606.1 | 2.38e-241 | 1 | 461 | 1 | 473 |
| QMW70533.1 | 2.38e-241 | 1 | 461 | 1 | 473 |
| QCP36276.1 | 6.93e-147 | 21 | 460 | 25 | 435 |
| BCD36607.1 | 2.26e-145 | 1 | 460 | 1 | 435 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5WQW_A | 1.18e-46 | 181 | 460 | 21 | 269 | X-raystructure of catalytic domain of autolysin from Clostridium perfringens [Clostridium perfringens str. 13] |
| 6FXO_A | 1.95e-19 | 217 | 458 | 57 | 241 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI7_A | 1.17e-12 | 253 | 441 | 79 | 209 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 3.90e-12 | 353 | 441 | 125 | 209 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 3.32e-19 | 217 | 458 | 699 | 877 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q99V41 | 2.81e-17 | 217 | 458 | 1061 | 1245 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q931U5 | 2.81e-17 | 217 | 458 | 1061 | 1245 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
| A7X0T9 | 2.82e-17 | 217 | 458 | 1068 | 1252 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu3 / ATCC 700698) OX=418127 GN=atl PE=3 SV=2 |
| Q6GI31 | 2.82e-17 | 217 | 458 | 1070 | 1254 | Bifunctional autolysin OS=Staphylococcus aureus (strain MRSA252) OX=282458 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
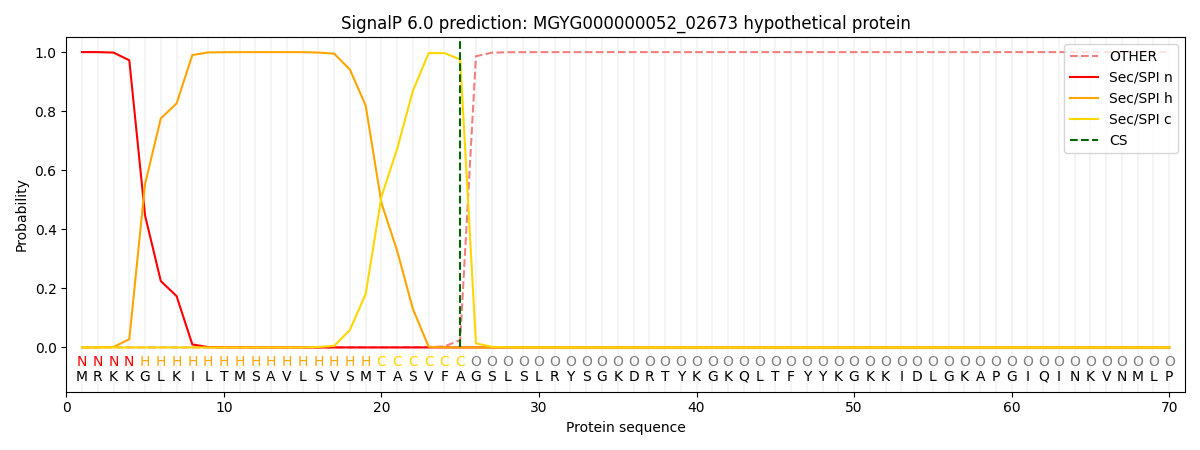
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000387 | 0.998789 | 0.000230 | 0.000198 | 0.000195 | 0.000169 |
