You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000054_01531
You are here: Home > Sequence: MGYG000000054_01531
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides acidifaciens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides acidifaciens | |||||||||||
| CAZyme ID | MGYG000000054_01531 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7050; End: 8570 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 197 | 484 | 1.6e-46 | 0.7661538461538462 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 2.62e-16 | 142 | 420 | 79 | 397 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 8.75e-09 | 210 | 452 | 48 | 276 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03003 | PLN03003 | 1.80e-07 | 209 | 490 | 106 | 372 | Probable polygalacturonase At3g15720 |
| PLN02793 | PLN02793 | 8.60e-06 | 242 | 314 | 180 | 254 | Probable polygalacturonase |
| PLN02218 | PLN02218 | 1.04e-05 | 242 | 497 | 218 | 422 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQA09746.1 | 0.0 | 1 | 506 | 1 | 506 |
| QQT79215.1 | 0.0 | 1 | 506 | 1 | 506 |
| ASW16392.1 | 0.0 | 1 | 506 | 1 | 506 |
| QRP58901.1 | 0.0 | 1 | 506 | 1 | 506 |
| SCM58771.1 | 4.09e-98 | 43 | 491 | 58 | 500 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLP_A | 1.35e-10 | 213 | 332 | 179 | 298 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 3JUR_A | 2.98e-09 | 238 | 485 | 189 | 415 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q949Z1 | 3.36e-10 | 241 | 421 | 212 | 363 | Polygalacturonase At1g48100 OS=Arabidopsis thaliana OX=3702 GN=At1g48100 PE=2 SV=1 |
| Q9LW07 | 3.86e-08 | 209 | 490 | 106 | 372 | Probable polygalacturonase At3g15720 OS=Arabidopsis thaliana OX=3702 GN=At3g15720 PE=3 SV=1 |
| A2QW66 | 8.10e-07 | 188 | 420 | 105 | 339 | Probable exopolygalacturonase A OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=pgxA PE=3 SV=1 |
| Q27UB2 | 8.10e-07 | 188 | 420 | 105 | 339 | Exopolygalacturonase A OS=Aspergillus niger OX=5061 GN=pgxA PE=3 SV=1 |
| Q873X6 | 3.33e-06 | 214 | 420 | 145 | 344 | Probable exopolygalacturonase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pgxB PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
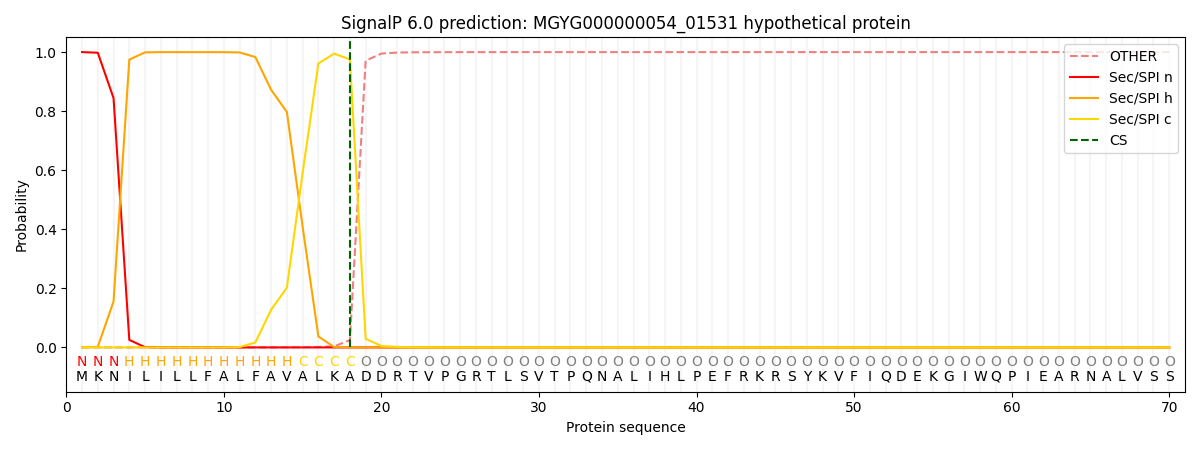
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000241 | 0.999158 | 0.000165 | 0.000151 | 0.000145 | 0.000129 |
