You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000054_01533
You are here: Home > Sequence: MGYG000000054_01533
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
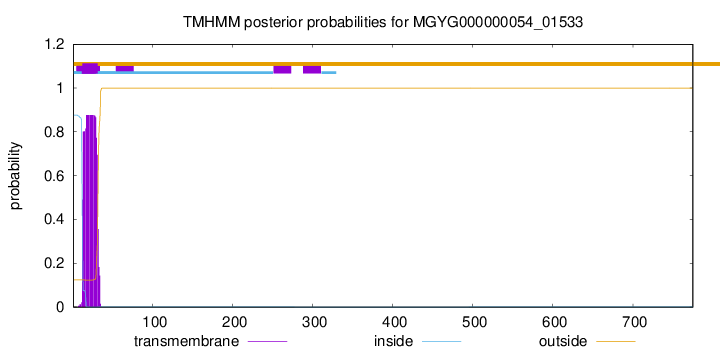
TMHMM annotations
Basic Information help
| Species | Bacteroides acidifaciens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides acidifaciens | |||||||||||
| CAZyme ID | MGYG000000054_01533 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 10472; End: 12799 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 473 | 754 | 1.2e-39 | 0.7446153846153846 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 7.41e-20 | 474 | 692 | 202 | 398 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam00295 | Glyco_hydro_28 | 7.62e-13 | 481 | 743 | 48 | 305 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| PLN03010 | PLN03010 | 2.14e-12 | 526 | 721 | 168 | 345 | polygalacturonase |
| PLN02793 | PLN02793 | 8.18e-11 | 528 | 721 | 190 | 365 | Probable polygalacturonase |
| PLN02218 | PLN02218 | 4.63e-09 | 562 | 753 | 241 | 410 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRP58902.1 | 0.0 | 1 | 775 | 1 | 775 |
| QQA09745.1 | 0.0 | 1 | 775 | 1 | 775 |
| QQT79214.1 | 0.0 | 1 | 775 | 1 | 775 |
| ASW16403.1 | 0.0 | 1 | 775 | 1 | 775 |
| BBL02017.1 | 3.75e-176 | 53 | 766 | 31 | 735 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1HG8_A | 3.60e-06 | 442 | 764 | 28 | 314 | Endopolygalacturonasefrom the phytopathogenic fungus Fusarium moniliforme [Fusarium verticillioides] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8RY29 | 3.05e-08 | 528 | 766 | 205 | 423 | Polygalacturonase ADPG2 OS=Arabidopsis thaliana OX=3702 GN=ADPG2 PE=2 SV=2 |
| Q0C7H7 | 9.37e-08 | 483 | 765 | 140 | 388 | Probable exopolygalacturonase B OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=pgxB PE=3 SV=1 |
| Q873X6 | 1.66e-07 | 483 | 749 | 146 | 380 | Probable exopolygalacturonase B OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=pgxB PE=2 SV=1 |
| Q4WBK6 | 2.26e-07 | 483 | 765 | 141 | 389 | Probable exopolygalacturonase B OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=pgxB PE=3 SV=2 |
| A1DAX2 | 2.26e-07 | 483 | 765 | 141 | 389 | Probable exopolygalacturonase B OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=pgxB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
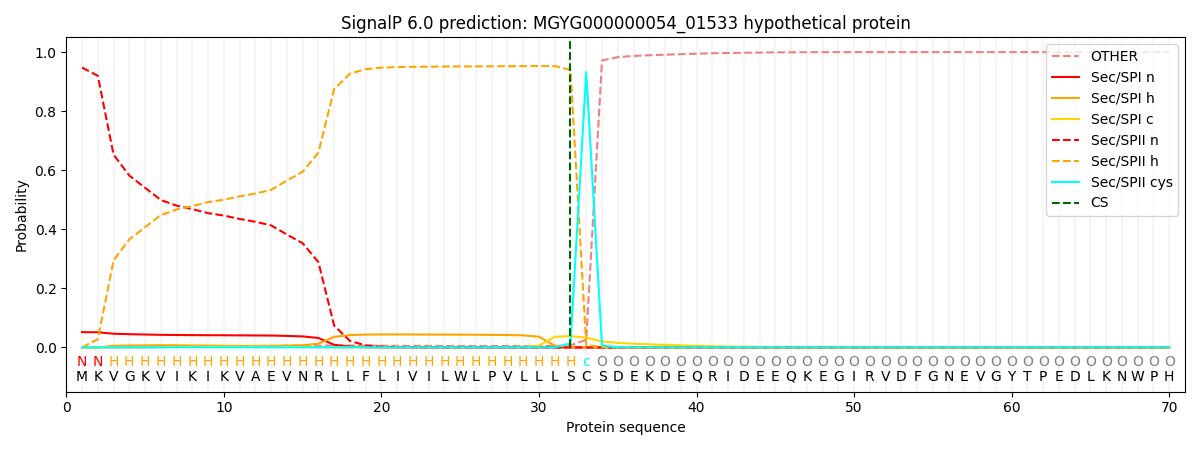
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000756 | 0.041163 | 0.957951 | 0.000032 | 0.000064 | 0.000033 |