You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000054_01774
You are here: Home > Sequence: MGYG000000054_01774
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides acidifaciens | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides acidifaciens | |||||||||||
| CAZyme ID | MGYG000000054_01774 | |||||||||||
| CAZy Family | PL6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25410; End: 27674 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL6 | 33 | 393 | 1.4e-131 | 0.9650537634408602 |
| PL6 | 450 | 656 | 1.9e-38 | 0.590778097982709 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd14251 | PL-6 | 0.0 | 33 | 393 | 6 | 366 | Polysaccharide Lyase Family 6. Polysaccharide Lyase Family 6 is a family of beta-helical polysaccharide lyases. Members include alginate lyase (EC 4.2.2.3) and chondroitinase B (EC 4.2.2.19). Chondroitinase B is an enzyme that only cleaves the beta-(1,4)-linkage of dermatan sulfate (DS), leading to 4,5-unsaturated dermatan sulfate disaccharides as the product. DS is a highly sulfated, unbranched polysaccharide belonging to a family of glycosaminoglycans (GAGs) composed of alternating hexosamine (gluco- or galactosamine) and uronic acid (D-glucuronic or L-iduronic acid) moieties. DS contains alternating 1,4-beta-D-galactosamine (GalNac) and 1,3-alpha-L-iduronic acid units. The related chondroitin sulfate (CS) contains alternating GalNac and 1,3-beta-D-glucuronic acid units. Alginate lyases (known as either mannuronate (EC 4.2.2.3) or guluronate lyases (EC 4.2.2.11) catalyze the degradation of alginate, a copolymer of alpha-L-guluronate and its C5 epimer beta-D-mannuronate. |
| pfam14592 | Chondroitinas_B | 2.15e-125 | 33 | 420 | 6 | 409 | Chondroitinase B. This family includes chondroitinases. These enzymes cleave the glycosaminoglycan dermatan sulfate. |
| pfam16318 | DUF4957 | 1.01e-10 | 470 | 595 | 1 | 119 | Domain of unknown function (DUF4957). This family consists of uncharacterized proteins around 150 residues in length and is mainly found in various Bacteroides and Prevotella species. The function of this protein is unknown. |
| pfam05048 | NosD | 7.28e-06 | 216 | 301 | 91 | 174 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
| pfam13229 | Beta_helix | 1.09e-05 | 549 | 686 | 33 | 144 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT26279.1 | 0.0 | 1 | 754 | 1 | 754 |
| QIU93442.1 | 0.0 | 1 | 751 | 1 | 751 |
| QNL39961.1 | 0.0 | 1 | 753 | 1 | 753 |
| QNT66846.1 | 1.06e-242 | 4 | 710 | 5 | 705 |
| QRQ49286.1 | 4.53e-235 | 27 | 751 | 23 | 755 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7DMK_A | 8.22e-240 | 31 | 744 | 17 | 733 | ChainA, BcAlyPL6 [Bacteroides clarus],7DMK_B Chain B, BcAlyPL6 [Bacteroides clarus],7DMK_C Chain C, BcAlyPL6 [Bacteroides clarus],7DMK_D Chain D, BcAlyPL6 [Bacteroides clarus] |
| 5GKD_A | 1.61e-139 | 38 | 679 | 15 | 678 | Structureof PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_B Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_C Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis],5GKD_D Structure of PL6 family alginate lyase AlyGC [Paraglaciecola chathamensis] |
| 5GKQ_A | 1.25e-138 | 38 | 679 | 15 | 678 | Structureof PL6 family alginate lyase AlyGC mutant-R241A [Paraglaciecola chathamensis S18K6],5GKQ_B Structure of PL6 family alginate lyase AlyGC mutant-R241A [Paraglaciecola chathamensis S18K6] |
| 7O77_A | 9.09e-129 | 31 | 679 | 7 | 678 | ChainA, Poly(Beta-D-mannuronate) lyase [Pseudoalteromonas atlantica T6c] |
| 7O7T_A | 9.09e-129 | 31 | 679 | 7 | 678 | ChainA, Poly(Beta-D-mannuronate) lyase [Pseudoalteromonas atlantica T6c] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q06365 | 2.85e-82 | 43 | 394 | 2 | 346 | Alginate lyase OS=Pseudomonas sp. (strain OS-ALG-9) OX=86038 GN=aly PE=3 SV=1 |
| Q46079 | 2.10e-26 | 38 | 340 | 38 | 349 | Chondroitinase-B OS=Pedobacter heparinus (strain ATCC 13125 / DSM 2366 / CIP 104194 / JCM 7457 / NBRC 12017 / NCIMB 9290 / NRRL B-14731 / HIM 762-3) OX=485917 GN=cslB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
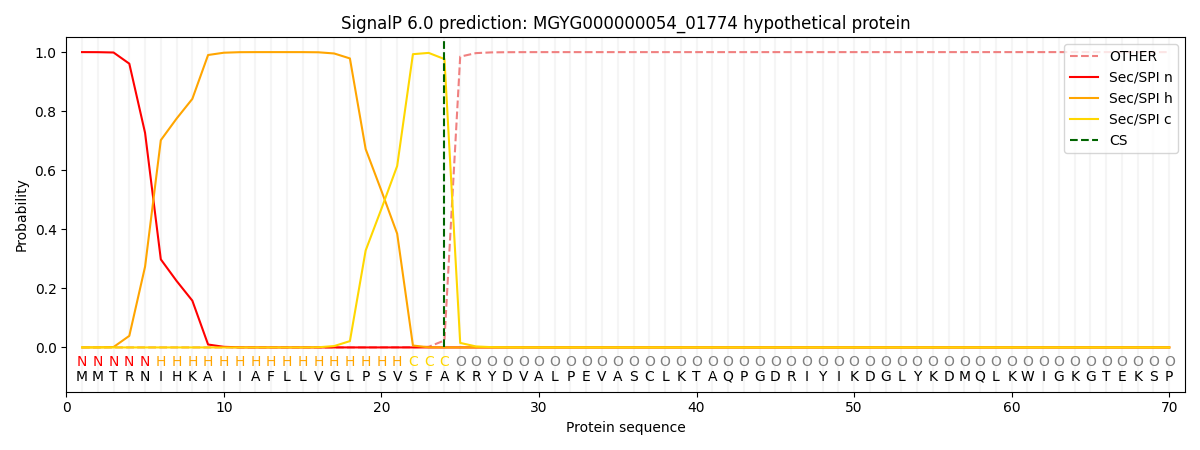
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000252 | 0.999048 | 0.000203 | 0.000158 | 0.000158 | 0.000145 |
