You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000055_02078
You are here: Home > Sequence: MGYG000000055_02078
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
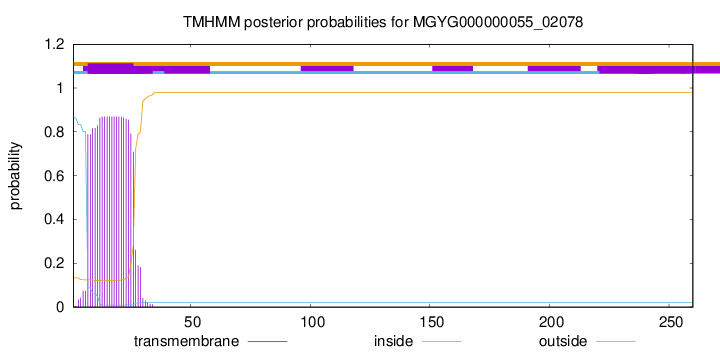
TMHMM annotations
Basic Information help
| Species | Muribaculum intestinale | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum intestinale | |||||||||||
| CAZyme ID | MGYG000000055_02078 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3284; End: 4066 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 76 | 260 | 1.6e-19 | 0.37538461538461537 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 1.12e-29 | 38 | 260 | 66 | 273 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN02218 | PLN02218 | 9.24e-06 | 6 | 162 | 2 | 175 | polygalacturonase ADPG |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB37320.1 | 4.19e-160 | 36 | 260 | 1 | 225 |
| ARE60831.1 | 4.19e-160 | 36 | 260 | 1 | 225 |
| QCD41743.1 | 5.92e-64 | 25 | 259 | 29 | 265 |
| QCP71949.1 | 1.72e-63 | 25 | 259 | 30 | 266 |
| QCD38261.1 | 1.72e-63 | 25 | 259 | 30 | 266 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 3.99e-26 | 36 | 259 | 15 | 224 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
| 5OLP_A | 9.03e-19 | 33 | 256 | 29 | 231 | Galacturonidase[Bacteroides thetaiotaomicron VPI-5482],5OLP_B Galacturonidase [Bacteroides thetaiotaomicron VPI-5482] |
| 2UVE_A | 5.25e-12 | 49 | 178 | 157 | 290 | Structureof Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVE_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase [Yersinia enterocolitica],2UVF_A Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica],2UVF_B Structure of Yersinia enterocolitica Family 28 Exopolygalacturonase in Complex with Digalaturonic Acid [Yersinia enterocolitica] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
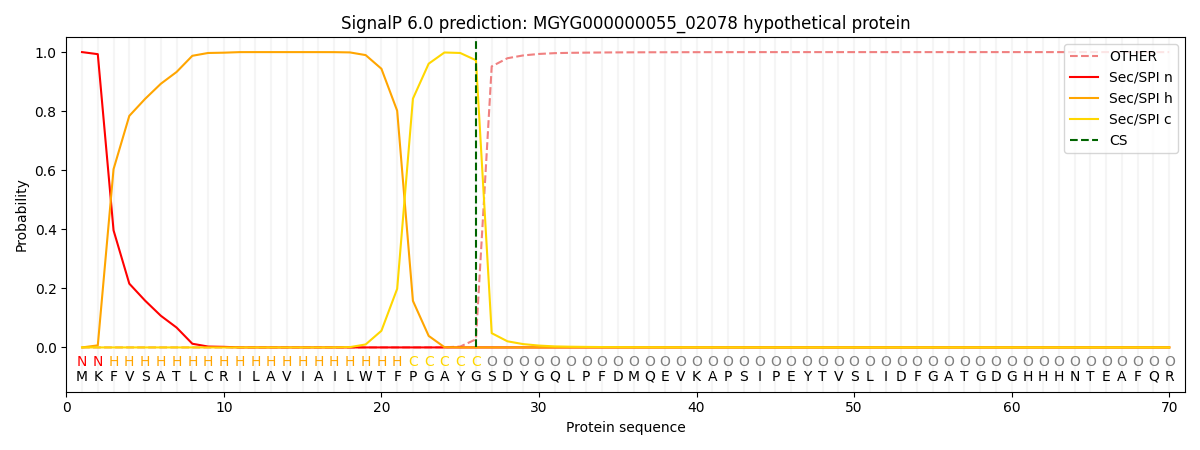
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000414 | 0.998884 | 0.000229 | 0.000158 | 0.000150 | 0.000149 |