You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000056_00921
You are here: Home > Sequence: MGYG000000056_00921
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Muribaculum sp002358615 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp002358615 | |||||||||||
| CAZyme ID | MGYG000000056_00921 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | Extracellular exo-alpha-L-arabinofuranosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 138940; End: 140934 Strand: + | |||||||||||
Full Sequence Download help
| MTLSKKLLTS LAVCAGLSLN AATNELVLQT NKPGAPIQPT MYGLFFEDIN YAADGGLYAE | 60 |
| LVKNRSFDFP QNLMGWTTFG GVELKNDGPF ERNPHYVRLT TAGHRDKHTG IENEGFFGIG | 120 |
| VKKGEEYRFS VWARNVGQAP AKIRVELVSP ESMGETQTIA SKEIEIKGDK WTKYTAVITP | 180 |
| SVTDAKSVLR VFLAKPDNAV VDLEHVSLFP VDTWNGHENG MRKDLAQKLA DLKPGIFRFP | 240 |
| GGCIVEGTDL PTRYQWKNTV GPVENRPVNE NRWHYTFPHR FFPDYYQSYG LGFYEYFLLS | 300 |
| EEIGSEPLPI LSVGLACQFQ NPGAEAHVPV DSLQPYIQDA LDLIEFANGG TDTKWGALRA | 360 |
| EMGHPAPFNL KYIGIGNEQW GPEYPEHLTP FLDAIRKAYP DIRIVGSSGP DSEGKQFDYL | 420 |
| WPEMKRLKAD LVDEHFYRPE SWFLAQGARY DNYDRKGPKV FAGEYACHGR GKKWNHFNAA | 480 |
| LLEAAFMTGL ERNADIVHMA TYAPLFAHVE GWQWRPDMIW YDNLSSIPTC SYFVQQLFAE | 540 |
| NKGTNVLQLT MDKKPLTGAE DQNGLFASAV YDKDADQYVV KVANTSDTPQ QLVLTFAGLK | 600 |
| KKESITDGTS YTLASKDLDL DNIERDGSMN LNIAPVKADI TVDGTKYDTT IAPNSFVVYR | 660 |
| FNKK | 664 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 198 | 662 | 1.3e-108 | 0.6857142857142857 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 2.83e-47 | 206 | 664 | 36 | 501 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| smart00813 | Alpha-L-AF_C | 3.01e-37 | 463 | 655 | 1 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06964 | Alpha-L-AF_C | 1.44e-33 | 463 | 655 | 1 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam02018 | CBM_4_9 | 1.13e-17 | 59 | 196 | 1 | 133 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCD36526.1 | 0.0 | 1 | 664 | 1 | 661 |
| QUT90799.1 | 0.0 | 7 | 663 | 7 | 659 |
| QIU95482.1 | 0.0 | 6 | 664 | 6 | 660 |
| ALJ58183.1 | 0.0 | 10 | 663 | 2 | 651 |
| QNL37257.1 | 0.0 | 7 | 664 | 7 | 660 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 1.53e-82 | 27 | 592 | 3 | 562 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 3S2C_A | 3.20e-16 | 218 | 655 | 46 | 475 | Structureof the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_B Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_C Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_D Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_E Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_F Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_G Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_H Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_I Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_J Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_K Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_L Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1] |
| 4ATW_A | 7.42e-16 | 218 | 655 | 46 | 475 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
| 3UG3_A | 8.09e-16 | 218 | 655 | 66 | 495 | Crystalstructure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG4_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG5_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima] |
| 2Y2W_A | 3.48e-13 | 218 | 407 | 89 | 246 | Elucidationof the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_B Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_C Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_D Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_E Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_F Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 1.06e-136 | 9 | 572 | 13 | 575 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 7.83e-106 | 30 | 656 | 48 | 650 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 6.08e-98 | 30 | 602 | 47 | 597 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| B8NKA3 | 3.10e-83 | 25 | 651 | 27 | 618 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfA PE=3 SV=2 |
| Q2U790 | 8.45e-83 | 25 | 598 | 27 | 575 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=abfA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
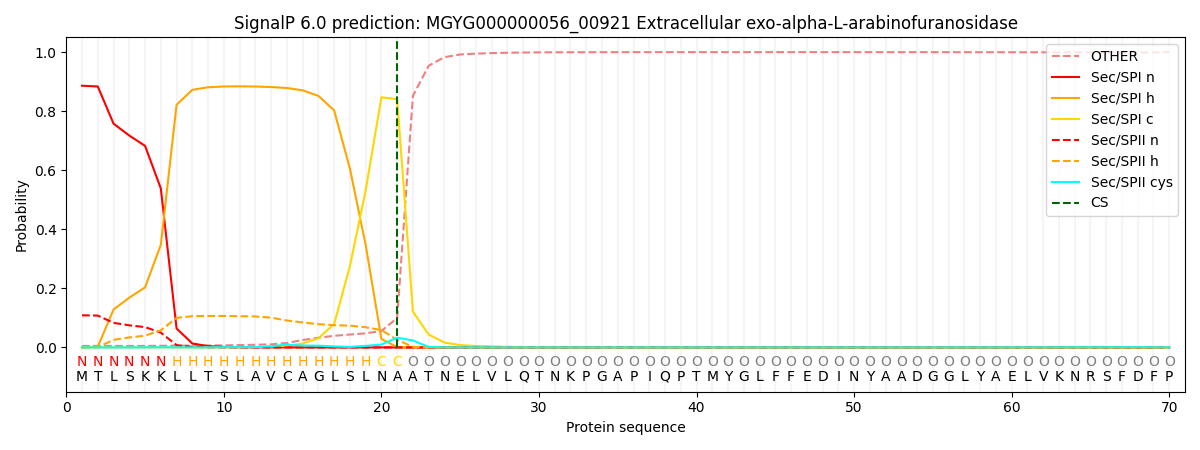
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.007240 | 0.877586 | 0.114208 | 0.000353 | 0.000287 | 0.000279 |
