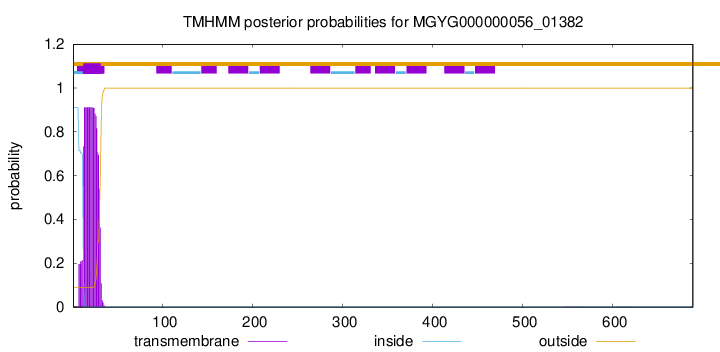
You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000056_01382
Basic Information
help
| Species |
Muribaculum sp002358615
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; Muribaculum; Muribaculum sp002358615
|
| CAZyme ID |
MGYG000000056_01382
|
| CAZy Family |
GH5 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000056 |
3324937 |
Isolate |
United Kingdom |
Europe |
|
| Gene Location |
Start: 104649;
End: 106718
Strand: -
|
| Family |
Start |
End |
Evalue |
family coverage |
| GH5 |
49 |
327 |
1e-114 |
0.9922779922779923 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
6.95e-20 |
43 |
278 |
1 |
201 |
Cellulase (glycosyl hydrolase family 5). |
| COG2730
|
BglC |
9.67e-05 |
94 |
222 |
85 |
212 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1G01_A
|
2.67e-08 |
42 |
291 |
29 |
248 |
AlkalineCellulase K Catalytic Domain [Bacillus sp. KSM-635],1G0C_A ALKALINE CELLULASE K CATALYTIC DOMAIN-CELLOBIOSE COMPLEX [Bacillus sp. KSM-635] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P19424
|
2.89e-07 |
42 |
291 |
249 |
468 |
Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
This protein is predicted as SP
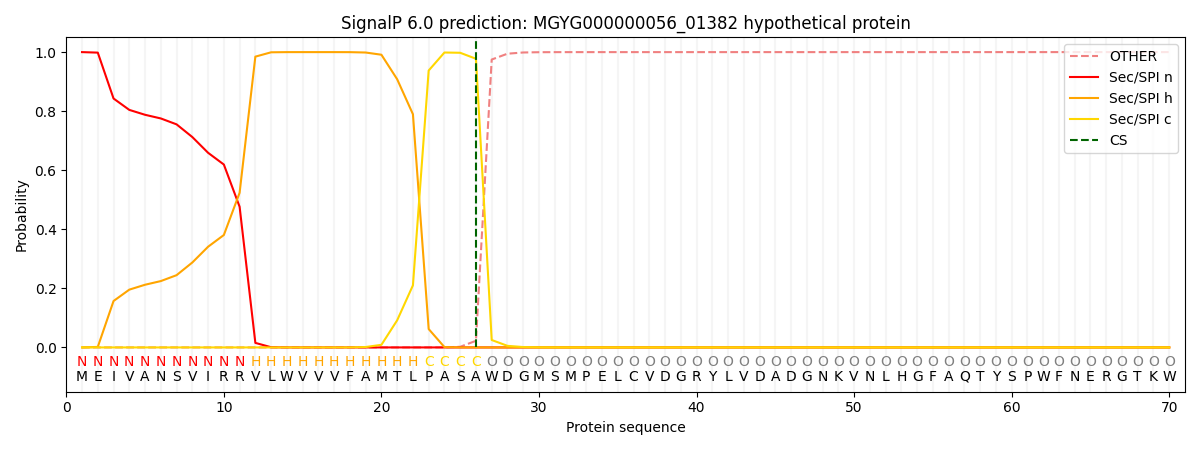
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000255
|
0.999109
|
0.000175
|
0.000158
|
0.000139
|
0.000135
|