You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000057_00146
You are here: Home > Sequence: MGYG000000057_00146
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides sp002491635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp002491635 | |||||||||||
| CAZyme ID | MGYG000000057_00146 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 170283; End: 172538 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 297 | 750 | 4.7e-107 | 0.6841269841269841 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 2.82e-48 | 312 | 750 | 42 | 498 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam06964 | Alpha-L-AF_C | 6.94e-48 | 559 | 744 | 1 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 7.71e-41 | 559 | 744 | 1 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam07944 | Glyco_hydro_127 | 4.98e-14 | 2 | 87 | 169 | 238 | Beta-L-arabinofuranosidase, GH127. One member of this family, from Bidobacterium longicum, UniProtKB:E8MGH8, has been characterized as an unusual beta-L-arabinofuranosidase enzyme, EC:3.2.1.185. It rleases l-arabinose from the l-arabinofuranose (Araf)-beta1,2-Araf disaccharide and also transglycosylates 1-alkanols with retention of the anomeric configuration. Terminal beta-l-arabinofuranosyl residues have been found in arabinogalactan proteins from a mumber of different plantt species. beta-l-Arabinofuranosyl linkages with 1-4 arabinofuranosides are also found in the sugar chains of extensin and solanaceous lectins, hydroxyproline (Hyp)2-rich glycoproteins that are widely observed in plant cell wall fractions. The critical residue for catalytic activity is Glu-338, in a ET/SCAS sequence context. |
| COG3533 | COG3533 | 3.29e-11 | 4 | 88 | 174 | 241 | Uncharacterized conserved protein, DUF1680 family [Function unknown]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMI82183.1 | 0.0 | 110 | 751 | 1 | 642 |
| QBJ20314.1 | 0.0 | 110 | 751 | 1 | 642 |
| QPH59624.1 | 0.0 | 110 | 751 | 1 | 642 |
| ALJ58902.1 | 0.0 | 110 | 750 | 1 | 642 |
| BBK85667.1 | 0.0 | 110 | 750 | 1 | 642 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 6.08e-90 | 131 | 681 | 1 | 557 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 6EX6_A | 2.68e-24 | 8 | 89 | 182 | 263 | TheGH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482],6EX6_B The GH127, Beta-arabinofuranosidase, BT3674 [Bacteroides thetaiotaomicron VPI-5482] |
| 2VRQ_A | 6.62e-14 | 318 | 693 | 50 | 436 | StructureOf An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_B Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_C Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus] |
| 6ZT6_A | 8.76e-14 | 318 | 693 | 50 | 436 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZTA_A | 1.53e-13 | 318 | 693 | 50 | 436 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 1.12e-134 | 131 | 644 | 37 | 545 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 1.68e-108 | 131 | 750 | 43 | 655 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 3.03e-102 | 121 | 750 | 30 | 652 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| B8NKA3 | 2.32e-78 | 128 | 751 | 21 | 629 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfA PE=3 SV=2 |
| Q2U790 | 2.32e-78 | 128 | 751 | 21 | 629 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=abfA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
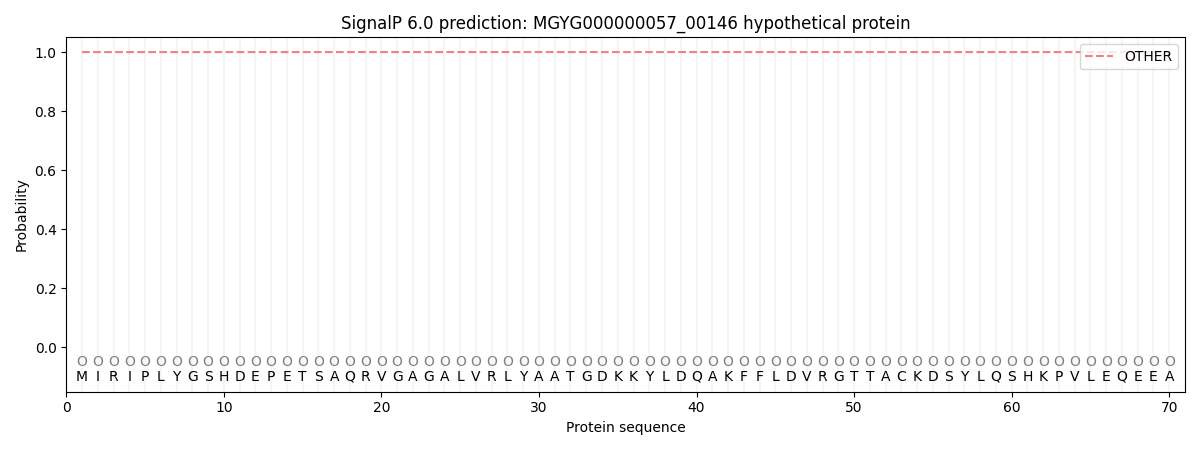
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000061 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
