You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000057_02669
You are here: Home > Sequence: MGYG000000057_02669
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
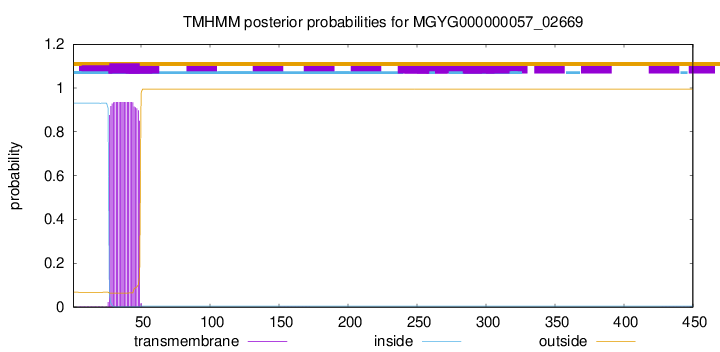
TMHMM annotations
Basic Information help
| Species | Bacteroides sp002491635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides sp002491635 | |||||||||||
| CAZyme ID | MGYG000000057_02669 | |||||||||||
| CAZy Family | GH158 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28430; End: 29782 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH158 | 59 | 189 | 2.2e-34 | 0.9772727272727273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 4.92e-11 | 57 | 220 | 292 | 468 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 6.15e-04 | 53 | 221 | 4 | 180 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QPH56445.1 | 3.61e-310 | 22 | 450 | 1 | 429 |
| QMI80893.1 | 3.61e-310 | 22 | 450 | 1 | 429 |
| QQA28931.1 | 4.91e-308 | 22 | 450 | 1 | 429 |
| BBK87700.1 | 4.91e-308 | 22 | 450 | 1 | 429 |
| QUT60461.1 | 4.91e-308 | 22 | 450 | 1 | 429 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PAL_A | 6.40e-293 | 46 | 450 | 3 | 407 | Bacteroidesuniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus [Bacteroides uniformis],6PAL_B Bacteroides uniformis endo-laminarinase BuGH158 from the beta(1,3)-glucan utilization locus [Bacteroides uniformis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NVK8 | 3.23e-06 | 48 | 189 | 100 | 239 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manA PE=3 SV=2 |
| Q2TXJ2 | 3.23e-06 | 48 | 189 | 100 | 239 | Probable mannan endo-1,4-beta-mannosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
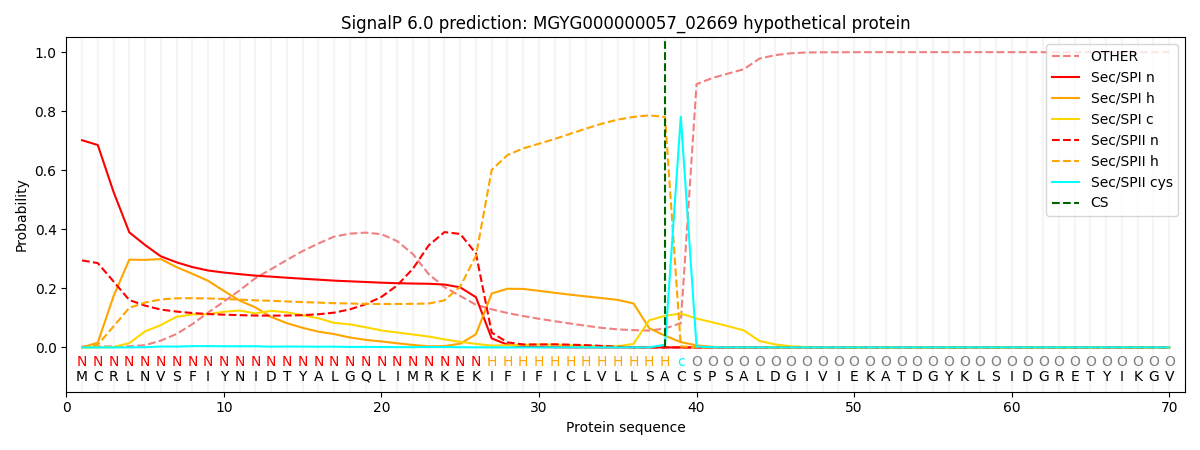
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002010 | 0.330903 | 0.666778 | 0.000129 | 0.000100 | 0.000072 |