You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000060_00409
You are here: Home > Sequence: MGYG000000060_00409
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
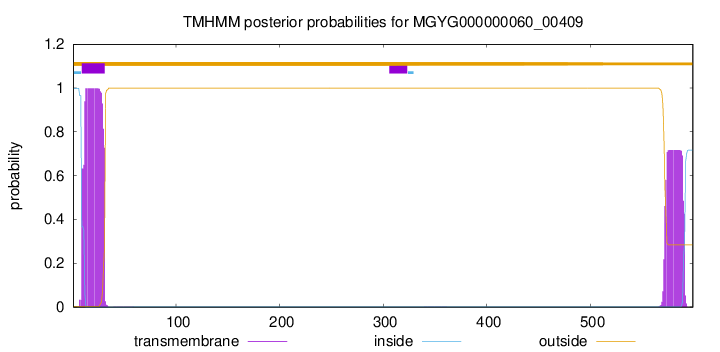
TMHMM annotations
Basic Information help
| Species | Lachnospira eligens_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnospira; Lachnospira eligens_A | |||||||||||
| CAZyme ID | MGYG000000060_00409 | |||||||||||
| CAZy Family | CE12 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 421828; End: 423627 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE12 | 41 | 260 | 3.3e-52 | 0.9904761904761905 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd01821 | Rhamnogalacturan_acetylesterase_like | 4.67e-50 | 39 | 260 | 1 | 198 | Rhamnogalacturan_acetylesterase_like subgroup of SGNH-hydrolases. Rhamnogalacturan acetylesterase removes acetyl esters from rhamnogalacturonan substrates, and renders them susceptible to degradation by rhamnogalacturonases. Rhamnogalacturonans are highly branched regions in pectic polysaccharides, consisting of repeating -(1,2)-L-Rha-(1,4)-D-GalUA disaccharide units, with many rhamnose residues substituted by neutral oligosaccharides such as arabinans, galactans and arabinogalactans. Extracellular enzymes participating in the degradation of plant cell wall polymers, such as Rhamnogalacturonan acetylesterase, would typically be found in saprophytic and plant pathogenic fungi and bacteria. |
| NF033839 | PspC_subgroup_2 | 2.73e-05 | 518 | 557 | 346 | 389 | pneumococcal surface protein PspC, LPXTG-anchored form. The pneumococcal surface protein PspC, as described in Streptococcus pneumoniae, is a repetitive and highly variable protein, recognized by a conserved N-terminal domain and also by genomic location. This form, subgroup 2, is anchored covalently after cleavage by sortase at a C-terminal LPXTG site. The other form, subgroup 1, has variable numbers of a choline-binding repeat in the C-terminal region, and is also known as choline-binding protein A. |
| NF033930 | pneumo_PspA | 3.81e-05 | 500 | 554 | 343 | 400 | pneumococcal surface protein A. The pneumococcal surface protein proteins, found in Streptococcus pneumoniae, are repetitive, with patterns of localized high sequence identity across pairs of proteins given different specific names that recombination may be presumed. This protein, PspA, has an N-terminal region that lacks a cross-wall-targeting YSIRK type extended signal peptide, in contrast to the closely related choline-binding protein CbpA which has a similar C-terminus but a YSIRK-containing region at the N-terminus. |
| pfam13472 | Lipase_GDSL_2 | 9.51e-05 | 43 | 212 | 1 | 150 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| NF033913 | fibronec_FbpA | 1.25e-04 | 511 | 565 | 200 | 266 | LPXTG-anchored fibronectin-binding protein FbpA. FbpA, a fibronectin-binding protein described in Streptococcus pyogenes, has a YSIRK-type (crosswall-targeting) signal peptide and a C-terminal LPXTG motif for covalent attachment to the cell wall. It is unrelated to the PavA-like protein from Streptococcus gordonii (see BlastRule NBR009716) that was given the identical name, so the phase LPXTG-anchored is added to the protein name for clarity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL50681.1 | 4.64e-245 | 1 | 517 | 1 | 523 |
| QAV09174.1 | 1.96e-219 | 31 | 513 | 1405 | 1888 |
| ACR72407.1 | 1.08e-173 | 41 | 505 | 4 | 472 |
| CBL00444.1 | 4.88e-121 | 17 | 510 | 810 | 1331 |
| QSI01052.1 | 7.37e-96 | 41 | 506 | 50 | 524 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1DEO_A | 6.42e-08 | 41 | 260 | 3 | 212 | RHAMNOGALACTURONANACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.55 A RESOLUTION WITH SO4 IN THE ACTIVE SITE [Aspergillus aculeatus],1DEX_A RHAMNOGALACTURONAN ACETYLESTERASE FROM ASPERGILLUS ACULEATUS AT 1.9 A RESOLUTION [Aspergillus aculeatus],1K7C_A Rhamnogalacturonan acetylesterase with seven N-linked carbohydrate residues distributed at two N-glycosylation sites refined at 1.12 A resolution [Aspergillus aculeatus],1PP4_A The crystal structure of rhamnogalacturonan acetylesterase in space group P3121 [Aspergillus aculeatus],1PP4_B The crystal structure of rhamnogalacturonan acetylesterase in space group P3121 [Aspergillus aculeatus] |
| 3C1U_A | 2.82e-07 | 41 | 260 | 3 | 212 | ChainA, Rhamnogalacturonan acetylesterase [Aspergillus aculeatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31523 | 8.96e-20 | 38 | 273 | 4 | 224 | Rhamnogalacturonan acetylesterase RhgT OS=Bacillus subtilis (strain 168) OX=224308 GN=rhgT PE=1 SV=1 |
| O31528 | 3.91e-10 | 41 | 273 | 5 | 214 | Probable rhamnogalacturonan acetylesterase YesY OS=Bacillus subtilis (strain 168) OX=224308 GN=yesY PE=1 SV=1 |
| P22751 | 2.36e-08 | 319 | 506 | 56 | 278 | Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
| Q00017 | 4.22e-07 | 41 | 260 | 20 | 229 | Rhamnogalacturonan acetylesterase OS=Aspergillus aculeatus OX=5053 GN=rha1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
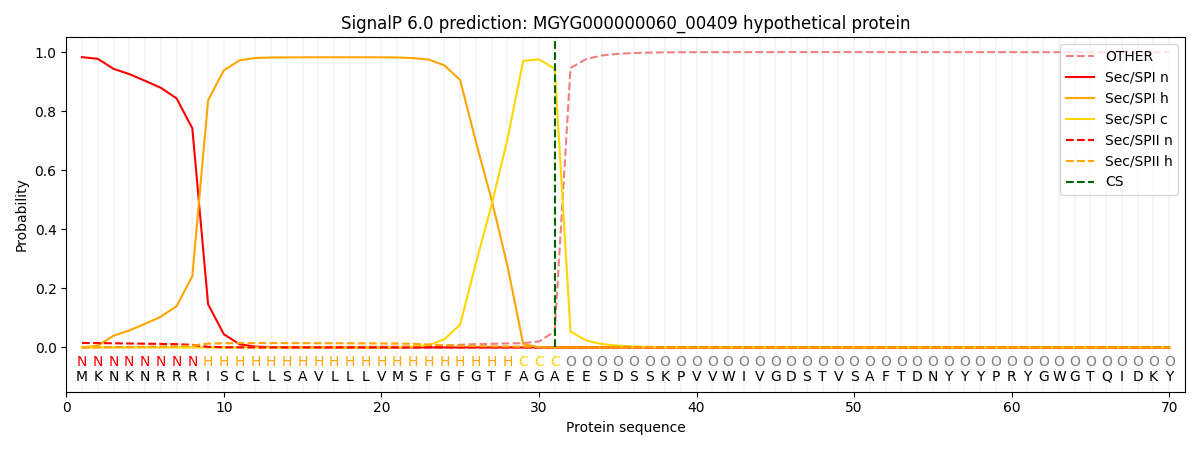
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002087 | 0.979467 | 0.016291 | 0.001646 | 0.000280 | 0.000208 |