You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000060_00510
You are here: Home > Sequence: MGYG000000060_00510
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
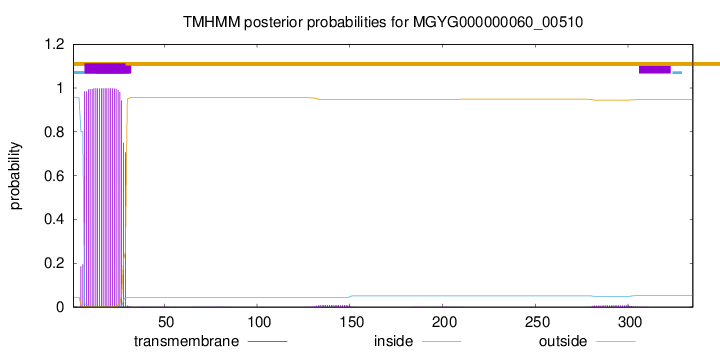
TMHMM annotations
Basic Information help
| Species | Lachnospira eligens_A | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnospira; Lachnospira eligens_A | |||||||||||
| CAZyme ID | MGYG000000060_00510 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 501945; End: 502952 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13702 | Lysozyme_like | 1.35e-61 | 48 | 204 | 1 | 165 | Lysozyme-like. |
| cd16891 | CwlT-like | 5.96e-54 | 54 | 205 | 1 | 151 | CwlT-like N-terminal lysozyme domain and similar domains. CwlT is a bifunctional cell wall hydrolase containing an N-terminal lysozyme domain and a C-terminal NlpC/P60 endopeptidase domain (gamma-d-D-glutamyl-L-diamino acid endopeptidase), and has been implicated in the spread of transposons. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
| pfam00877 | NLPC_P60 | 3.86e-34 | 226 | 332 | 1 | 105 | NlpC/P60 family. The function of this domain is unknown. It is found in several lipoproteins. |
| PRK13914 | PRK13914 | 5.30e-31 | 214 | 333 | 366 | 481 | invasion associated endopeptidase. |
| COG0791 | Spr | 1.63e-24 | 214 | 330 | 75 | 196 | Cell wall-associated hydrolase, NlpC family [Cell wall/membrane/envelope biogenesis]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCK80082.1 | 1.37e-245 | 1 | 335 | 1 | 335 |
| QNM11264.1 | 1.53e-242 | 1 | 335 | 1 | 335 |
| QNM13610.1 | 1.53e-242 | 1 | 335 | 1 | 335 |
| QFX79249.1 | 1.53e-242 | 1 | 335 | 1 | 335 |
| AQP39780.1 | 1.53e-242 | 1 | 335 | 1 | 335 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4HPE_A | 3.08e-214 | 29 | 335 | 2 | 308 | ChainA, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_B Chain B, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_C Chain C, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_D Chain D, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_E Chain E, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630],4HPE_F Chain F, Putative cell wall hydrolase Tn916-like,CTn1-Orf17 [Clostridioides difficile 630] |
| 4FDY_A | 5.39e-137 | 30 | 333 | 5 | 310 | ChainA, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50],4FDY_B Chain B, Similar to lipoprotein, NLP/P60 family [Staphylococcus aureus subsp. aureus Mu50] |
| 7CFL_A | 3.27e-15 | 214 | 333 | 14 | 137 | ChainA, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_B Chain B, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_C Chain C, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile],7CFL_D Chain D, Putative cell wall hydrolase phosphatase-associated protein [Clostridioides difficile] |
| 6BIQ_A | 1.26e-11 | 210 | 320 | 154 | 264 | Structureof NlpC2 from Trichomonas vaginalis [Trichomonas vaginalis],6BIQ_B Structure of NlpC2 from Trichomonas vaginalis [Trichomonas vaginalis],6BIQ_C Structure of NlpC2 from Trichomonas vaginalis [Trichomonas vaginalis],6BIQ_D Structure of NlpC2 from Trichomonas vaginalis [Trichomonas vaginalis] |
| 2EVR_A | 1.64e-11 | 229 | 333 | 126 | 237 | ChainA, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102],2FG0_A Chain A, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102],2FG0_B Chain B, COG0791: Cell wall-associated hydrolases (invasion-associated proteins) [Nostoc punctiforme PCC 73102] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P96645 | 2.11e-73 | 14 | 334 | 15 | 329 | Probable endopeptidase YddH OS=Bacillus subtilis (strain 168) OX=224308 GN=yddH PE=3 SV=1 |
| P21171 | 2.42e-23 | 217 | 333 | 372 | 484 | Probable endopeptidase p60 OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=iap PE=1 SV=2 |
| Q01839 | 1.36e-22 | 220 | 333 | 415 | 524 | Probable endopeptidase p60 OS=Listeria welshimeri OX=1643 GN=iap PE=3 SV=1 |
| O34636 | 1.76e-22 | 54 | 214 | 55 | 214 | Uncharacterized membrane protein YocA OS=Bacillus subtilis (strain 168) OX=224308 GN=yocA PE=4 SV=1 |
| Q01836 | 4.75e-22 | 220 | 333 | 358 | 467 | Probable endopeptidase p60 OS=Listeria innocua serovar 6a (strain ATCC BAA-680 / CLIP 11262) OX=272626 GN=iap PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
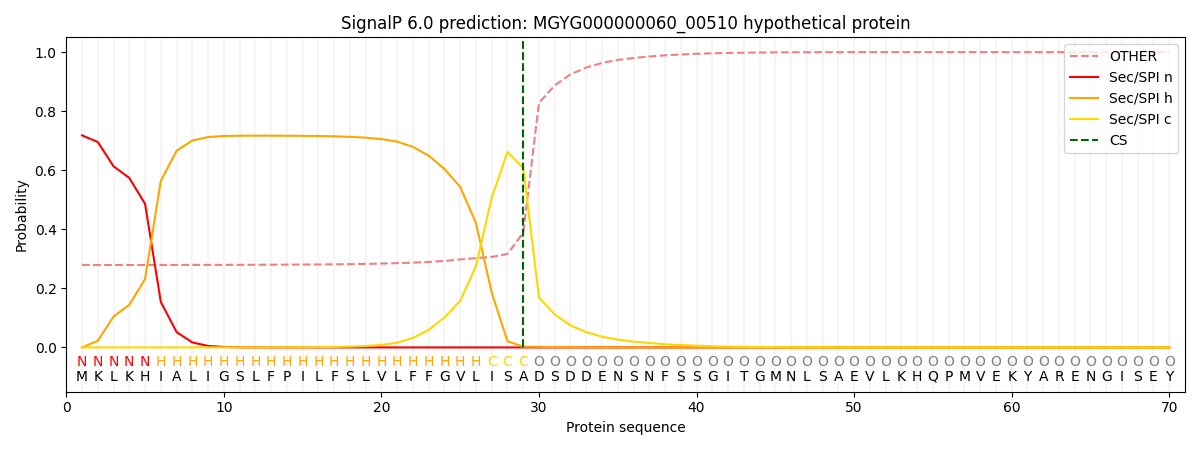
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.287974 | 0.707182 | 0.003810 | 0.000316 | 0.000276 | 0.000416 |