You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000062_02054
You are here: Home > Sequence: MGYG000000062_02054
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Intestinibacter bartlettii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Peptostreptococcaceae; Intestinibacter; Intestinibacter bartlettii | |||||||||||
| CAZyme ID | MGYG000000062_02054 | |||||||||||
| CAZy Family | CBM51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 94819; End: 100854 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM51 | 1621 | 1760 | 3.4e-38 | 0.9925373134328358 |
| CBM51 | 1398 | 1536 | 1.1e-37 | 0.9850746268656716 |
| CBM51 | 883 | 1018 | 5.7e-34 | 0.9850746268656716 |
| CBM32 | 1131 | 1254 | 7.4e-16 | 0.9112903225806451 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam08305 | NPCBM | 6.76e-48 | 1620 | 1761 | 2 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 1.85e-45 | 880 | 1019 | 1 | 136 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| pfam08305 | NPCBM | 2.17e-44 | 1396 | 1537 | 1 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| smart00776 | NPCBM | 9.76e-41 | 1620 | 1761 | 4 | 145 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
| smart00776 | NPCBM | 1.00e-35 | 1395 | 1537 | 2 | 144 | This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AQM59432.1 | 0.0 | 71 | 2008 | 497 | 2443 |
| QJS19250.1 | 0.0 | 88 | 2006 | 515 | 2286 |
| AMN35289.1 | 1.39e-296 | 71 | 1994 | 491 | 2141 |
| AWE07805.1 | 1.49e-296 | 88 | 2010 | 287 | 1925 |
| CEJ73532.1 | 5.28e-295 | 70 | 1318 | 265 | 1492 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JS4_A | 5.76e-141 | 321 | 1765 | 47 | 980 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JRM_A | 1.51e-127 | 1541 | 2007 | 4 | 462 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JRL_A | 3.10e-127 | 1541 | 2007 | 26 | 484 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 7JFS_A | 8.19e-111 | 88 | 801 | 268 | 981 | ChainA, F5/8 type C domain protein [Clostridium perfringens ATCC 13124] |
| 5KDJ_A | 3.02e-93 | 231 | 879 | 24 | 672 | ZmpBmetallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124],5KDJ_B ZmpB metallopeptidase from Clostridium perfringens [Clostridium perfringens ATCC 13124] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P42548 | 3.96e-06 | 1337 | 1637 | 396 | 641 | Uncharacterized 81.3 kDa protein OS=Acholeplasma phage L2 OX=46014 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
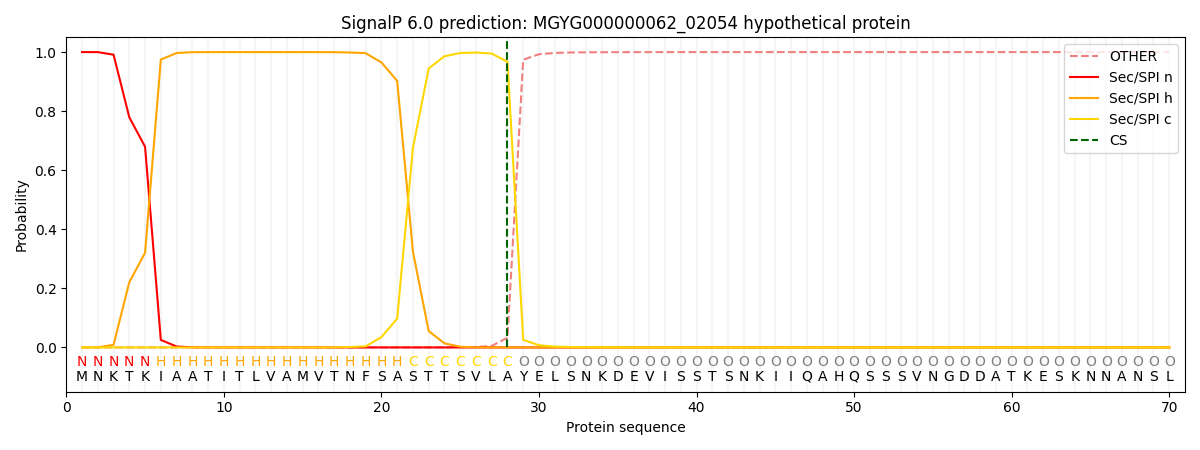
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000260 | 0.999099 | 0.000162 | 0.000174 | 0.000157 | 0.000146 |
