You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000066_00404
You are here: Home > Sequence: MGYG000000066_00404
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
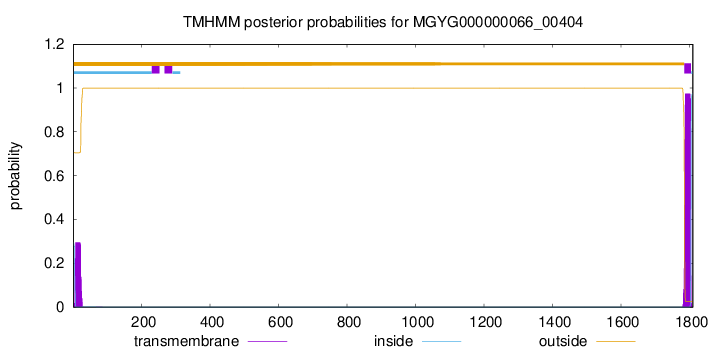
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp900066755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp900066755 | |||||||||||
| CAZyme ID | MGYG000000066_00404 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 164112; End: 169544 Strand: - | |||||||||||
Full Sequence Download help
| MRRLLTIILG VTLLLGTFPA TVRAGNNDVS SGTEKEFYVS PDGNDSNAGT KEEPFKTVER | 60 |
| ARVEVAKFNQ NMTADIVVYL MDGVYYQDKS LTFGTGDSGT NGYKVRYEAY QDAKPEISGG | 120 |
| KQLTGTWELH DAEKNIYKIP VEEGLNFRQL YVNGEKGIRA RTGEPGIFDG SSRIQGADRL | 180 |
| DENGNVIPEW WRDQGNAAAK VQAADGRVIV PADGRISEDM DGLRQIELHI FTAWSENILR | 240 |
| VDTLEKIDHY DCQAEAQHTG KEVEWDGACY RLKIQNPEAV RIFNRPHPGL DNYAGGPHYA | 300 |
| FYYENAYQYI DEDTEWYLDT DKNMLYYKAP ADMDMSTASA VAPVLDEVIV LEGTLDNPVH | 360 |
| DIVFKGLTVE HSTWLQPSEE GLVGGQACQY VTYGIFANND IGVKRADAGI RIENAAHIRL | 420 |
| DGNHIRFMGA TGILLASGTN EVTLINNTVE ETAGNGIEVG KFAVDENTDY HTAYNPADLR | 480 |
| EVCTNDRILN NTIHTIGTQY EGAVGIGAGY VQGITIGNNT IYDCPYSGIS VGYGWTSTPN | 540 |
| PMKRNNISRN EIYHVNQIVC DGGAIYTLSN QEPDSMILEN YMHDNMLPEG ADYGANGIYM | 600 |
| DEQTSGYTVM DNVLVYSYGV SQHITGPNNM QTNYTFWGKD TDNGYDAFVT DKVKSVMENA | 660 |
| GVQDPYDEEE LFKPVLDGAD YDPYYGILNL TGRKFGSDAG TVSIETESGT VMLTSDDIKS | 720 |
| WSNGEISFVP PQNVASGDMV TVTTKDKITS AAYPVGVLAD SVTELLKEDF DGQEEGRLND | 780 |
| SDWSVSVADK ASIVSGESGN YLELKGNDPN LNVSKLDQNG TVLAFGNNVT QFDFQFPQEL | 840 |
| SGYTGLYNTL RVTDQKTEYT ANIRPAYQTK LAIELKGQEE VKDNDKDLET GVWYTCRTMV | 900 |
| YENMIYLSVF KKGDPEPDGW GLKKKMADEV KKDCLLNFSF YDPSGRSVWI DNITVSKYSD | 960 |
| EKSKPVISGT DYDPVHQMIT ITGQGFSDDT GSVTYKTADG EGTVGGEKIL LWTNTRINLI | 1020 |
| KPEDAVDGVV SVTTSKNITS NQDVILELPV PAVFYEEDFN VLTQEQFEAQ YDVSVAEKAN | 1080 |
| IAEETGTDGN RMLLLKGNDP NLEVDLLNDE LQHVVYGENI TTFDFQFPQG SYSWYGMYNV | 1140 |
| LRDTGASVFR VNIKPAQNVK YYLDLKDIGE LPLNGKEIKN AVWYTCKTMV KGNSLYLKAW | 1200 |
| EKSAQEPSNW ELVREMPNQT GKECRLNFSY YDPDGRPLYI DNIKVEVLDK ADVERRLNGI | 1260 |
| EIKAEPDKNI YYIDDEFDPS GLKVNAVYND GSSQELADGT YTLSGFESTA EGEKVITVSY | 1320 |
| RGFTAEFTVQ VTQKPVITEI KITQQPYKTI YEQGEELDLS GLVVKAVYDN TTEEVISLSD | 1380 |
| CTVEGFSSDT EGTKTVQLSY QEFHAEFTVE VKSPAAGEKT LTGIRIDQQP DKTEYLIGED | 1440 |
| LNLEGLLVTA IYEDNSEEPL AGGTYSVTGF DSETEGTKTV TVRYKEQTAD FTVQVKAAEL | 1500 |
| PEKILNMIRI DQLPEKTEYF IGEELETAGL SVTAVYEDTS EEVLDPDAYT LTGFDSETEG | 1560 |
| TKTITVTYQD KTAEFQVEVK KEAEPTPTPS PTPTPTQTPT PTPSPTPTPT PTPTPSPTPT | 1620 |
| PDPTPVPGQA VISSEGIKYY ILQSVLPENK RNEANAVLVQ NQEYQGLAGQ YPDKNRIFID | 1680 |
| ISVLDQNGGQ IHETQGALKI TVPYPDHSDS TKDFIIFHVK NDGQAERITT FEKTDSGLRI | 1740 |
| TADSFSTYVI GWKDAAKQES HTDNGGGESH DSDNNAQGAQ TGDSAAPVIW LLCCGAALSL | 1800 |
| ILILRKRSRI | 1810 |
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 36 | 614 | 4.6e-143 | 0.9886148007590133 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 9.00e-13 | 408 | 571 | 1 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 3.55e-11 | 431 | 626 | 1 | 146 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam07523 | Big_3 | 2.19e-10 | 1514 | 1579 | 2 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam07523 | Big_3 | 3.93e-10 | 1429 | 1495 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
| pfam07523 | Big_3 | 1.19e-08 | 1265 | 1331 | 1 | 67 | Bacterial Ig-like domain (group 3). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL27132.1 | 9.09e-109 | 25 | 756 | 18 | 692 |
| ACX75367.1 | 9.09e-109 | 25 | 756 | 18 | 692 |
| QDN75355.1 | 8.18e-97 | 37 | 638 | 37 | 589 |
| QDN84965.1 | 4.14e-95 | 37 | 638 | 37 | 589 |
| QDO17263.1 | 4.14e-95 | 37 | 638 | 37 | 589 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 8.89e-61 | 36 | 633 | 26 | 600 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A401ETL2 | 1.69e-07 | 1401 | 1573 | 1269 | 1445 | Exo-beta-1,6-galactobiohydrolase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=bl1,6Gal PE=1 SV=1 |
| P94576 | 5.03e-07 | 13 | 186 | 8 | 178 | Uncharacterized protein YwoF OS=Bacillus subtilis (strain 168) OX=224308 GN=ywoF PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000288 | 0.999040 | 0.000179 | 0.000183 | 0.000167 | 0.000147 |