You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000066_00874
You are here: Home > Sequence: MGYG000000066_00874
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
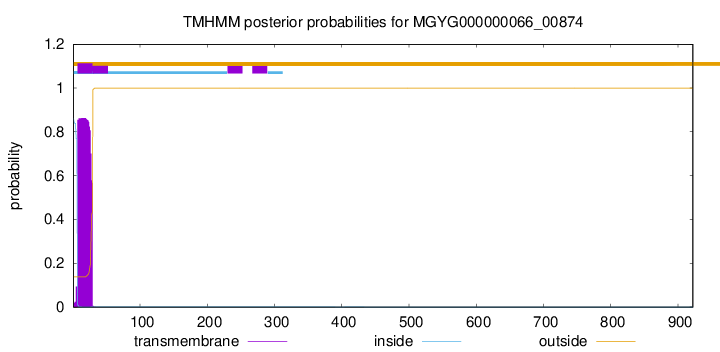
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp900066755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp900066755 | |||||||||||
| CAZyme ID | MGYG000000066_00874 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 183712; End: 186480 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 39 | 558 | 2.1e-130 | 0.9886148007590133 |
| CBM32 | 655 | 765 | 9.4e-19 | 0.9032258064516129 |
| CBM22 | 775 | 890 | 6.7e-18 | 0.8931297709923665 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 2.31e-13 | 648 | 763 | 1 | 122 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02018 | CBM_4_9 | 1.42e-09 | 774 | 905 | 2 | 132 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| cd00057 | FA58C | 2.41e-05 | 650 | 738 | 14 | 102 | Substituted updates: Jan 31, 2002 |
| pfam13229 | Beta_helix | 4.27e-04 | 354 | 554 | 8 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJD84702.1 | 3.65e-199 | 37 | 768 | 33 | 773 |
| QGQ98753.1 | 7.21e-128 | 35 | 777 | 34 | 772 |
| BAV33387.1 | 2.12e-100 | 58 | 630 | 44 | 578 |
| BBH24245.1 | 2.53e-98 | 6 | 701 | 7 | 689 |
| QGQ94344.1 | 8.58e-96 | 31 | 630 | 35 | 601 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQP_A | 1.70e-41 | 43 | 559 | 33 | 586 | Glycosidehydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_B Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_C Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_D Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_E Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_F Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_G Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron],5MQP_H Glycoside hydrolase BT_1002 [Bacteroides thetaiotaomicron] |
| 2RV9_A | 7.15e-08 | 643 | 768 | 7 | 135 | Solutionstructure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
| 4ZXE_A | 7.31e-08 | 643 | 768 | 8 | 136 | X-raycrystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_B X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis],4ZXE_C X-ray crystal structure of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5. [Paenibacillus fukuinensis] |
| 4ZY9_A | 2.51e-07 | 643 | 768 | 8 | 136 | X-raycrystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis],4ZY9_B X-ray crystal structure of selenomethionine-labelled V110M mutant of chitosan-binding module 1 derived from chitosanase/glucanase from Paenibacillus sp. IK-5 [Paenibacillus fukuinensis] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000338 | 0.999006 | 0.000191 | 0.000151 | 0.000141 | 0.000140 |