You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000066_02476
You are here: Home > Sequence: MGYG000000066_02476
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
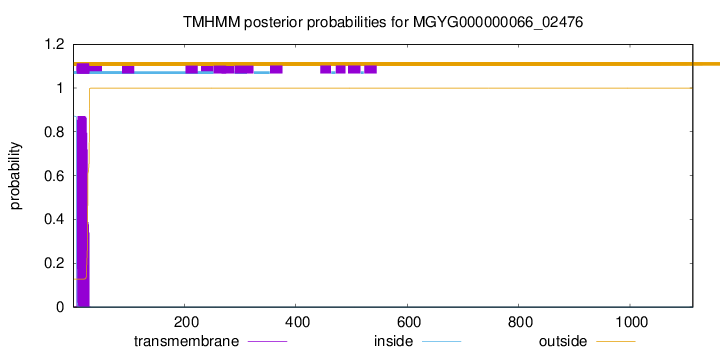
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp900066755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp900066755 | |||||||||||
| CAZyme ID | MGYG000000066_02476 | |||||||||||
| CAZy Family | GH136 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 79493; End: 82834 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH136 | 48 | 526 | 9.1e-129 | 0.9918533604887984 |
| CBM16 | 668 | 777 | 1.7e-24 | 0.9568965517241379 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| NF033675 | NTTRR-F1 | 1.24e-09 | 668 | 723 | 4 | 61 | NTTRR-F1 domain. NTTRR-F1 (N-terminal To Repetitive Region - Firmicutes 1) is a homology domain found strictly as the N-terminal non-repetitive region of otherwise highly repetitive proteins of various Firmicutes. The repetitive region that follows typically is collagen-like, with every third residue a glycine. |
| pfam02018 | CBM_4_9 | 5.73e-09 | 668 | 778 | 2 | 127 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| pfam13229 | Beta_helix | 1.21e-06 | 249 | 397 | 8 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 6.77e-06 | 175 | 369 | 3 | 155 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 7.66e-04 | 133 | 319 | 3 | 136 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUH27719.1 | 5.11e-117 | 49 | 793 | 36 | 781 |
| BCA49028.1 | 8.71e-97 | 48 | 566 | 42 | 563 |
| QGN59088.1 | 2.25e-92 | 40 | 796 | 33 | 655 |
| QYX75179.1 | 1.05e-90 | 49 | 796 | 40 | 791 |
| QGQ95108.1 | 2.12e-90 | 10 | 560 | 13 | 547 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7V6M_A | 2.93e-53 | 49 | 528 | 11 | 578 | ChainA, Fibronectin type III domain-containing protein [Tyzzerella nexilis] |
| 6N1B_A | 3.73e-47 | 803 | 1111 | 24 | 403 | Crystalstructure of an N-acetylgalactosamine deacetylase from F. plautii in complex with blood group B trisaccharide [Flavonifractor plautii] |
| 6N1A_A | 7.50e-46 | 803 | 1111 | 24 | 403 | Crystalstructure of an N-acetylgalactosamine deacetylase from F. plautii [Flavonifractor plautii] |
| 5GQC_A | 1.28e-38 | 49 | 528 | 20 | 598 | Crystalstructure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_C Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_D Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_E Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_F Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_G Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQC_H Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, ligand-free form [Bifidobacterium longum subsp. longum],5GQF_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQF_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, lacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_A Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum],5GQG_B Crystal structure of lacto-N-biosidase LnbX from Bifidobacterium longum subsp. longum, galacto-N-biose complex [Bifidobacterium longum subsp. longum] |
| 7V6I_A | 6.42e-38 | 49 | 530 | 16 | 612 | ChainA, Lacto-N-biosidase [Bifidobacterium saguini DSM 23967] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR4 | 8.02e-45 | 803 | 1111 | 30 | 409 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| P0DTR5 | 1.59e-16 | 532 | 665 | 706 | 828 | A type blood alpha-D-galactosamine galactosaminidase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| Q9ZG90 | 1.13e-14 | 661 | 799 | 283 | 422 | Keratan-sulfate endo-1,4-beta-galactosidase OS=Sphingobacterium multivorum OX=28454 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000266 | 0.999015 | 0.000204 | 0.000185 | 0.000163 | 0.000149 |