You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000066_04291
You are here: Home > Sequence: MGYG000000066_04291
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
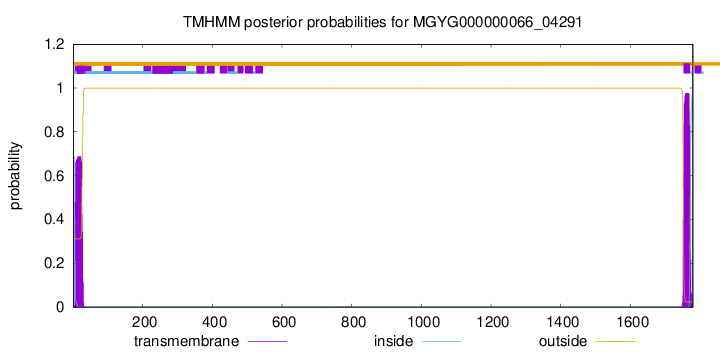
TMHMM annotations
Basic Information help
| Species | GCA-900066755 sp900066755 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; GCA-900066755; GCA-900066755 sp900066755 | |||||||||||
| CAZyme ID | MGYG000000066_04291 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24819; End: 30161 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 454 | 1084 | 1.6e-48 | 0.8282548476454293 |
| CBM32 | 301 | 435 | 1.8e-19 | 0.9354838709677419 |
| CBM32 | 148 | 275 | 3e-16 | 0.9193548387096774 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.90e-17 | 300 | 434 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 3.80e-16 | 145 | 274 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 1.13e-09 | 171 | 277 | 36 | 142 | Substituted updates: Jan 31, 2002 |
| cd00057 | FA58C | 2.13e-07 | 326 | 414 | 36 | 127 | Substituted updates: Jan 31, 2002 |
| smart00231 | FA58C | 2.88e-06 | 326 | 437 | 35 | 137 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 1.05e-203 | 36 | 1131 | 40 | 1090 |
| AWS40658.1 | 9.07e-156 | 30 | 1126 | 31 | 926 |
| QXE33659.1 | 1.72e-144 | 29 | 1126 | 55 | 952 |
| BCB76148.1 | 8.22e-143 | 298 | 1132 | 87 | 882 |
| QKW17790.1 | 1.33e-82 | 453 | 1108 | 135 | 759 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GDB_A | 2.33e-07 | 125 | 259 | 786 | 924 | Crystalstructure of Spr0440 glycoside hydrolase domain, Endo-D from Streptococcus pneumoniae R6 [Streptococcus pneumoniae R6] |
| 2XQX_A | 5.00e-07 | 161 | 259 | 32 | 130 | Structureof the family 32 carbohydrate-binding module from Streptococcus pneumoniae EndoD [Streptococcus pneumoniae TIGR4],2XQX_B Structure of the family 32 carbohydrate-binding module from Streptococcus pneumoniae EndoD [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| E8MGH9 | 7.69e-08 | 1128 | 1325 | 1662 | 1842 | Beta-L-arabinobiosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=hypBA2 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
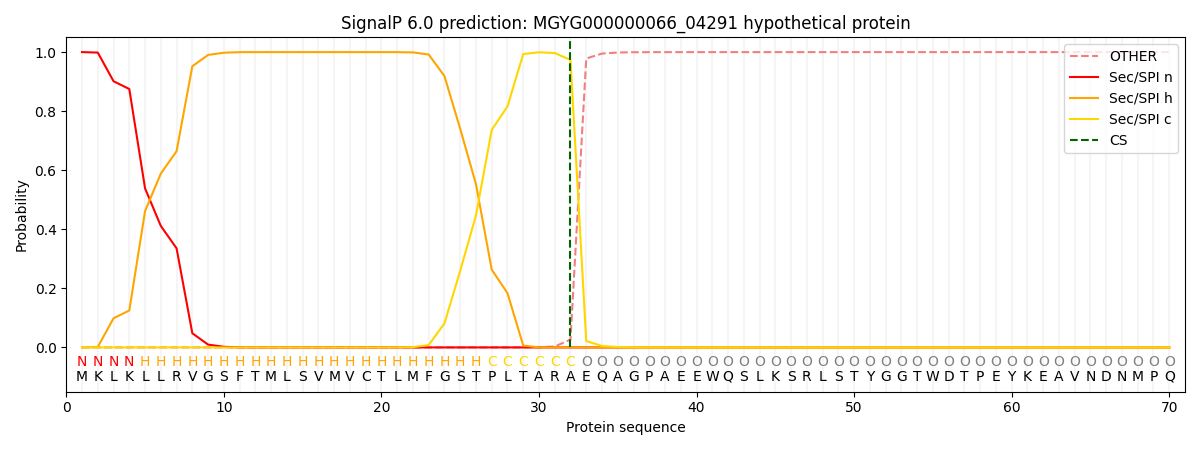
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000227 | 0.999082 | 0.000196 | 0.000180 | 0.000152 | 0.000139 |