You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000067_00326
You are here: Home > Sequence: MGYG000000067_00326
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
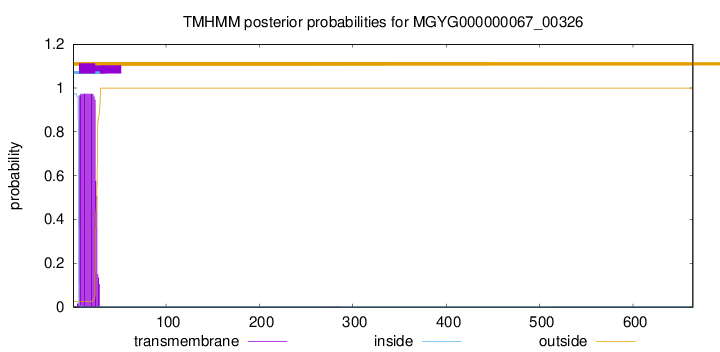
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium_B sp900066765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; Lachnoclostridium_B sp900066765 | |||||||||||
| CAZyme ID | MGYG000000067_00326 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 331654; End: 333648 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.53e-18 | 195 | 386 | 39 | 242 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 7.56e-10 | 264 | 321 | 8 | 75 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| pfam00395 | SLH | 7.11e-09 | 544 | 586 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 4.62e-08 | 526 | 664 | 562 | 696 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| NF033190 | inl_like_NEAT_1 | 4.70e-08 | 482 | 591 | 641 | 749 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QOV20091.1 | 8.55e-222 | 4 | 660 | 4 | 661 |
| QEK17141.1 | 2.72e-160 | 1 | 482 | 1 | 496 |
| AWY97555.1 | 1.27e-147 | 3 | 483 | 2 | 490 |
| QNM11023.1 | 1.82e-108 | 72 | 508 | 139 | 579 |
| QPS13527.1 | 1.42e-100 | 39 | 482 | 76 | 517 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 7.92e-33 | 111 | 375 | 57 | 284 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 1.24e-30 | 111 | 375 | 425 | 652 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 1.50e-30 | 111 | 375 | 469 | 696 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
| P19424 | 3.03e-16 | 484 | 659 | 41 | 214 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| P38536 | 2.39e-14 | 349 | 664 | 1553 | 1860 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 8.04e-14 | 484 | 664 | 908 | 1086 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
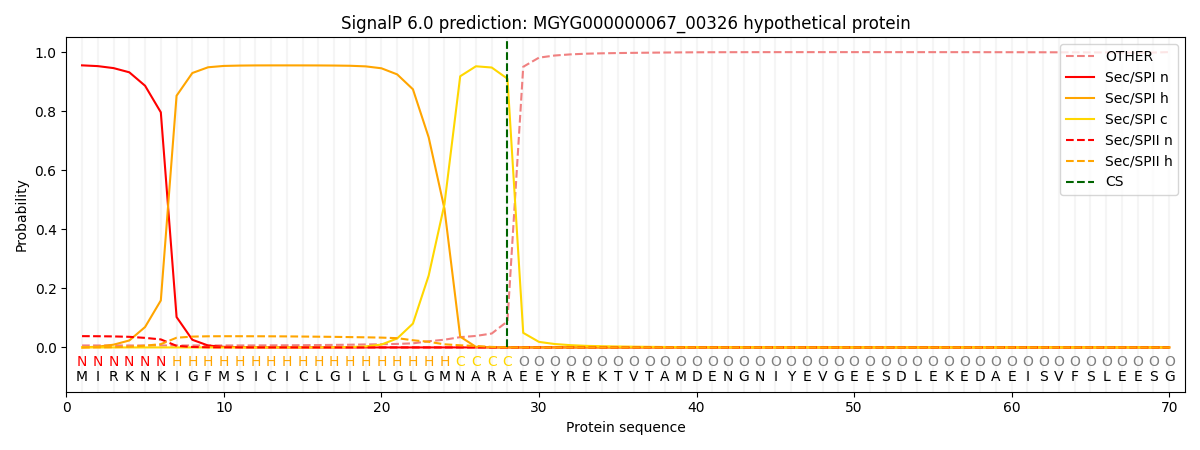
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.011808 | 0.945888 | 0.041109 | 0.000468 | 0.000348 | 0.000334 |