You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000067_00648
You are here: Home > Sequence: MGYG000000067_00648
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium_B sp900066765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; Lachnoclostridium_B sp900066765 | |||||||||||
| CAZyme ID | MGYG000000067_00648 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 184139; End: 186898 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE17 | 598 | 758 | 1.6e-41 | 0.9878787878787879 |
| CE1 | 297 | 535 | 2.7e-38 | 0.9691629955947136 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2382 | Fes | 7.32e-22 | 287 | 540 | 66 | 298 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam00756 | Esterase | 1.72e-18 | 297 | 479 | 1 | 175 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| cd00229 | SGNH_hydrolase | 1.02e-14 | 596 | 768 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| COG2819 | YbbA | 9.57e-14 | 291 | 449 | 2 | 171 | Predicted hydrolase of the alpha/beta superfamily [General function prediction only]. |
| pfam13472 | Lipase_GDSL_2 | 2.37e-13 | 599 | 760 | 2 | 176 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADU21606.1 | 1.77e-144 | 294 | 919 | 99 | 982 |
| CBL17231.1 | 4.34e-95 | 257 | 539 | 801 | 1073 |
| CBL17903.1 | 8.54e-88 | 256 | 539 | 685 | 954 |
| ADU21987.1 | 2.81e-74 | 257 | 539 | 965 | 1232 |
| CAB93667.1 | 1.10e-62 | 290 | 539 | 546 | 792 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5CXU_A | 1.61e-63 | 275 | 538 | 11 | 270 | Structureof the CE1 ferulic acid esterase AmCE1/Fae1A, from the anaerobic fungi Anaeromyces mucronatus in the absence of substrate [Anaeromyces mucronatus],5CXX_A Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus],5CXX_B Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus],5CXX_C Structure of a CE1 ferulic acid esterase, AmCE1/Fae1A, from Anaeromyces mucronatus in complex with Ferulic acid [Anaeromyces mucronatus] |
| 6HH9_A | 9.41e-40 | 563 | 787 | 2 | 231 | Crystalstructure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82],6HH9_B Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82],6HH9_C Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82],6HH9_D Crystal structure of a two-domain esterase (CEX) active on acetylated mannans co-crystallized with mannopentaose [Roseburia intestinalis L1-82] |
| 6HFZ_A | 1.45e-38 | 563 | 787 | 2 | 231 | Crystalstructure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82],6HFZ_B Crystal structure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82],6HFZ_C Crystal structure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82],6HFZ_D Crystal structure of a two-domain esterase (CEX) active on acetylated mannans [Roseburia intestinalis L1-82] |
| 1JJF_A | 2.24e-36 | 275 | 535 | 17 | 255 | ChainA, Endo-1,4-beta-xylanase Z [Acetivibrio thermocellus] |
| 1JT2_A | 5.62e-36 | 275 | 535 | 17 | 255 | ChainA, PROTEIN (ENDO-1,4-BETA-XYLANASE Z) [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P10478 | 1.38e-32 | 275 | 535 | 36 | 274 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| P19424 | 1.78e-18 | 1 | 220 | 1 | 219 | Endoglucanase OS=Bacillus sp. (strain KSM-635) OX=1415 PE=1 SV=1 |
| D5EY13 | 1.63e-14 | 285 | 444 | 495 | 650 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| P38536 | 1.91e-14 | 27 | 216 | 1675 | 1856 | Amylopullulanase OS=Thermoanaerobacterium thermosulfurigenes OX=33950 GN=amyB PE=3 SV=2 |
| P38535 | 1.08e-13 | 33 | 216 | 903 | 1082 | Exoglucanase XynX OS=Acetivibrio thermocellus OX=1515 GN=xynX PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
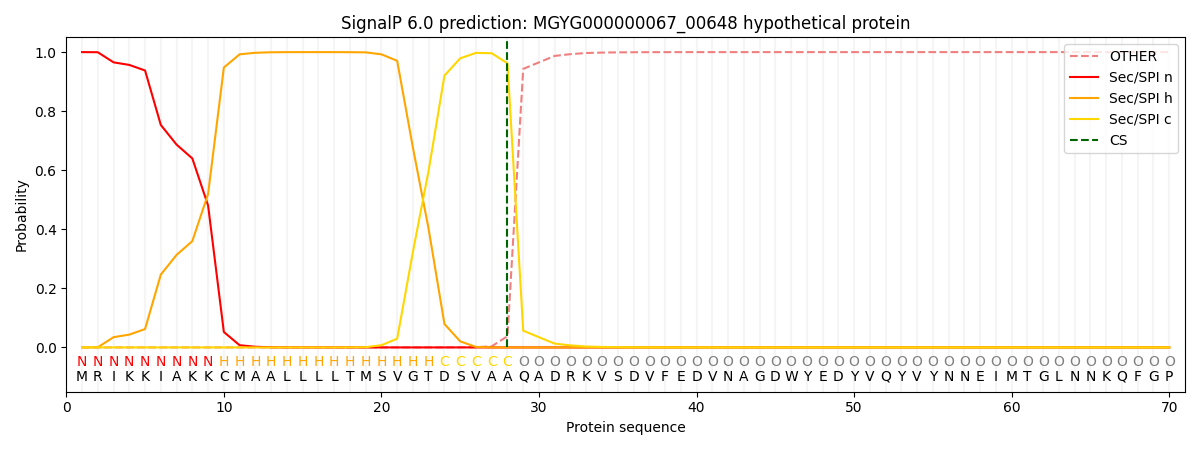
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000228 | 0.999101 | 0.000201 | 0.000162 | 0.000145 | 0.000136 |
