You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000067_01261
You are here: Home > Sequence: MGYG000000067_01261
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
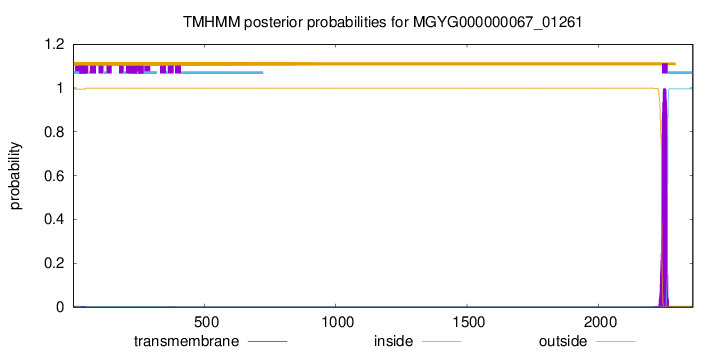
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium_B sp900066765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; Lachnoclostridium_B sp900066765 | |||||||||||
| CAZyme ID | MGYG000000067_01261 | |||||||||||
| CAZy Family | CBM13 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 148713; End: 155798 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM13 | 1292 | 1397 | 3.1e-18 | 0.5425531914893617 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3209 | RhsA | 3.65e-21 | 1609 | 2228 | 43 | 674 | Uncharacterized conserved protein RhsA, contains 28 RHS repeats [General function prediction only]. |
| TIGR03696 | Rhs_assc_core | 1.99e-18 | 2122 | 2209 | 1 | 76 | RHS repeat-associated core domain. This model represents a conserved unique core sequence shared by large numbers of proteins. It is occasional in the Archaea Methanosarcina barkeri) but common in bacteria and eukaryotes. Most fall into two large classes. One class consists of long proteins in which two classes of repeats are abundant: an FG-GAP repeat (pfam01839) class, and an RHS repeat (pfam05593) or YD repeat (TIGR01643). This class includes secreted bacterial insecticidal toxins and intercellular signalling proteins such as the teneurins in animals. The other class consists of uncharacterized proteins shorter than 400 amino acids, where this core domain of about 75 amino acids tends to occur in the N-terminal half. Over twenty such proteins are found in Pseudomonas putida alone; little sequence similarity or repeat structure is found among these proteins outside the region modeled by this domain. |
| NF033679 | DNRLRE_dom | 5.26e-18 | 327 | 488 | 2 | 156 | DNRLRE domain. The DNRLRE domain, with a length of about 160 amino acids, appears typically in large, repetitive surface proteins of bacteria and archaea, sometimes repeated several times. It occurs, notably, three times in the C-terminal region of the enzyme disaggregatase from the archaeal species Methanosarcina mazei, each time with the motif DNRLRE, for which the domain is named. Archaeal proteins within this family are described particularly well by the currently more narrowly defined Pfam model, PF06848. Note that the catalytic region of disaggregatase, in the N-terminal portion of the protein, is modeled by a different HMM, PF08480. |
| pfam14200 | RicinB_lectin_2 | 3.38e-17 | 1295 | 1370 | 15 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 3.61e-14 | 1328 | 1419 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM08600.1 | 0.0 | 11 | 2210 | 80 | 2309 |
| QRT30109.1 | 0.0 | 12 | 2230 | 3 | 2170 |
| QEI31369.1 | 0.0 | 12 | 2213 | 3 | 2158 |
| QHB23864.1 | 0.0 | 12 | 2213 | 3 | 2158 |
| QRT48460.1 | 0.0 | 12 | 2209 | 3 | 2324 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6H6G_A | 2.57e-08 | 1950 | 2209 | 1879 | 2156 | CrystalStructure of TcdB2-TccC3 without hypervariable C-terminal region [Photorhabdus luminescens] |
| 6SUP_A | 2.57e-08 | 1950 | 2209 | 1837 | 2114 | CrystalStructure of TcdB2-TccC3-Cdc42 [Photorhabdus luminescens] |
| 4O9X_A | 2.58e-08 | 1950 | 2209 | 1913 | 2190 | CrystalStructure of TcdB2-TccC3 [Photorhabdus luminescens] |
| 6H6E_F | 2.61e-08 | 1950 | 2209 | 1874 | 2151 | PTC3holotoxin complex from Photorhabdus luminecens in prepore state (TcdA1, TcdB2, TccC3) [Photorhabdus luminescens] |
| 6SUF_F | 2.61e-08 | 1950 | 2209 | 1879 | 2156 | Structureof Photorhabdus luminescens Tc holotoxin pore [Photorhabdus luminescens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| G4NYJ6 | 8.07e-31 | 1950 | 2218 | 1938 | 2196 | tRNA3(Ser)-specific nuclease WapA OS=Bacillus spizizenii (strain DSM 15029 / JCM 12233 / NBRC 101239 / NRRL B-23049 / TU-B-10) OX=1052585 GN=wapA PE=1 SV=1 |
| Q07833 | 1.81e-30 | 1950 | 2218 | 1938 | 2196 | tRNA nuclease WapA OS=Bacillus subtilis (strain 168) OX=224308 GN=wapA PE=1 SV=2 |
| D4G3R4 | 1.52e-27 | 1987 | 2218 | 1981 | 2209 | tRNA(Glu)-specific nuclease WapA OS=Bacillus subtilis subsp. natto (strain BEST195) OX=645657 GN=wapA PE=1 SV=2 |
| P0DUH5 | 3.16e-13 | 1885 | 2216 | 950 | 1294 | Double-stranded DNA deaminase toxin A OS=Burkholderia cenocepacia (strain H111) OX=1055524 GN=dddA PE=1 SV=1 |
| P77759 | 1.65e-07 | 2106 | 2209 | 1 | 93 | Putative uncharacterized protein YlbH OS=Escherichia coli (strain K12) OX=83333 GN=ylbH PE=5 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
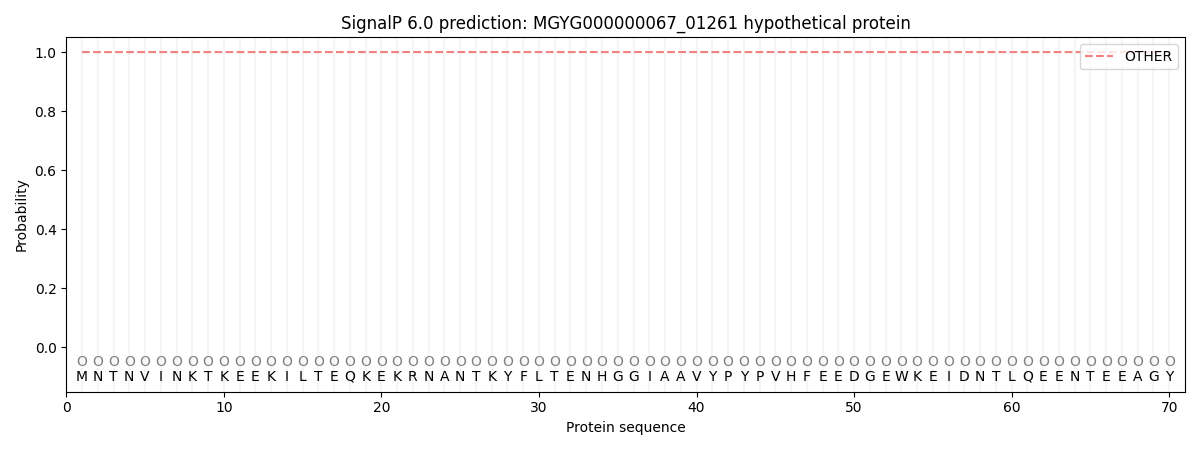
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000049 | 0.000004 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |