You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000067_02215
You are here: Home > Sequence: MGYG000000067_02215
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
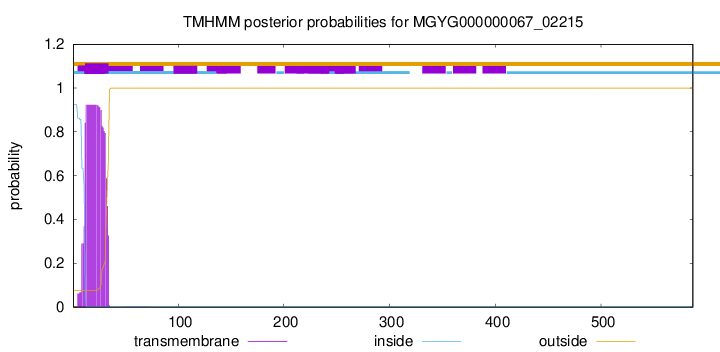
TMHMM annotations
Basic Information help
| Species | Lachnoclostridium_B sp900066765 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Lachnoclostridium_B; Lachnoclostridium_B sp900066765 | |||||||||||
| CAZyme ID | MGYG000000067_02215 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61454; End: 63217 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 248 | 580 | 2.5e-95 | 0.976897689768977 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00331 | Glyco_hydro_10 | 1.26e-105 | 248 | 580 | 2 | 307 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 3.72e-96 | 288 | 580 | 1 | 262 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.90e-63 | 279 | 578 | 58 | 334 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| NF033190 | inl_like_NEAT_1 | 4.91e-11 | 40 | 177 | 580 | 715 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
| pfam00395 | SLH | 4.12e-08 | 102 | 144 | 1 | 42 | S-layer homology domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CUH92237.1 | 2.20e-108 | 240 | 587 | 256 | 605 |
| SIP63107.1 | 2.78e-99 | 230 | 587 | 30 | 391 |
| BAA21516.2 | 1.34e-82 | 247 | 582 | 199 | 542 |
| ANV77297.1 | 1.34e-82 | 247 | 582 | 199 | 542 |
| ABN53059.1 | 1.34e-82 | 247 | 582 | 199 | 542 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FHE_A | 7.60e-66 | 250 | 582 | 16 | 339 | Highlyactive enzymes by automated modular backbone assembly and sequence design [synthetic construct] |
| 6D5C_A | 1.15e-63 | 249 | 582 | 25 | 348 | Structureof Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_B Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii],6D5C_C Structure of Caldicellulosiruptor danielii GH10 module of glycoside hydrolase WP_045175321 [Caldicellulosiruptor danielii] |
| 2W5F_A | 4.59e-63 | 224 | 578 | 164 | 521 | ChainA, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus],2W5F_B Chain B, ENDO-1,4-BETA-XYLANASE Y [Acetivibrio thermocellus] |
| 7NL2_A | 3.50e-62 | 247 | 582 | 12 | 338 | ChainA, Beta-xylanase [Pseudothermotoga thermarum DSM 5069],7NL2_B Chain B, Beta-xylanase [Pseudothermotoga thermarum DSM 5069] |
| 1N82_A | 7.41e-62 | 247 | 582 | 8 | 329 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P26223 | 5.01e-77 | 247 | 587 | 3 | 344 | Endo-1,4-beta-xylanase B OS=Butyrivibrio fibrisolvens OX=831 GN=xynB PE=3 SV=1 |
| P23557 | 2.81e-64 | 288 | 579 | 1 | 296 | Putative endo-1,4-beta-xylanase OS=Caldicellulosiruptor saccharolyticus OX=44001 PE=3 SV=1 |
| P51584 | 2.37e-60 | 224 | 578 | 175 | 532 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| P10474 | 5.06e-60 | 249 | 582 | 47 | 370 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| P45703 | 3.20e-58 | 247 | 579 | 8 | 325 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
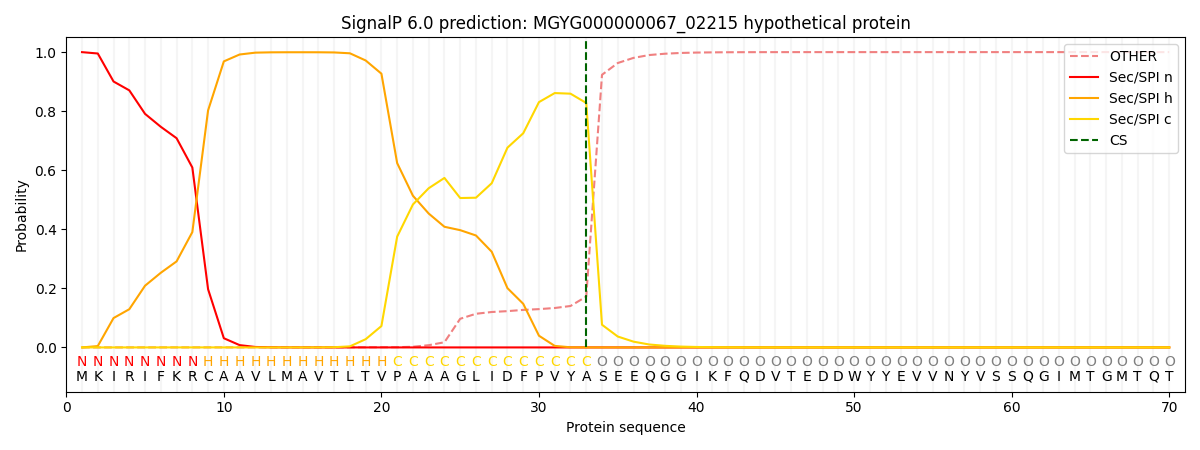
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000537 | 0.998656 | 0.000194 | 0.000250 | 0.000175 | 0.000150 |