You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000068_01646
You are here: Home > Sequence: MGYG000000068_01646
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Agathobaculum sp900291975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Butyricicoccaceae; Agathobaculum; Agathobaculum sp900291975 | |||||||||||
| CAZyme ID | MGYG000000068_01646 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 84796; End: 86739 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01832 | Glucosaminidase | 7.43e-10 | 514 | 580 | 2 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| cd20614 | CYPBJ-4-like | 0.002 | 180 | 278 | 208 | 325 | cytochrome P450 BJ-4 homolog and similar cytochrome P450s. This group is composed of mostly uncharacterized proteins including Sinorhizobium fredii CYPBJ-4 homolog. It belongs to the large cytochrome P450 (P450, CYP) superfamily of heme-containing proteins that catalyze a variety of oxidative reactions of a large number of structurally different endogenous and exogenous compounds in organisms from all major domains of life. CYPs bind their diverse ligands in a buried, hydrophobic active site, which is accessed through a substrate access channel formed by two flexible helices and their connecting loop. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AFC63685.1 | 1.98e-70 | 480 | 644 | 202 | 366 |
| QMW80001.1 | 4.30e-68 | 479 | 645 | 171 | 337 |
| QIB57222.1 | 4.30e-68 | 479 | 645 | 171 | 337 |
| QUO23193.1 | 1.78e-65 | 485 | 645 | 2 | 162 |
| ADL53623.1 | 1.79e-63 | 479 | 645 | 194 | 360 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O51481 | 3.92e-22 | 475 | 647 | 35 | 198 | Uncharacterized protein BB_0531 OS=Borreliella burgdorferi (strain ATCC 35210 / DSM 4680 / CIP 102532 / B31) OX=224326 GN=BB_0531 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
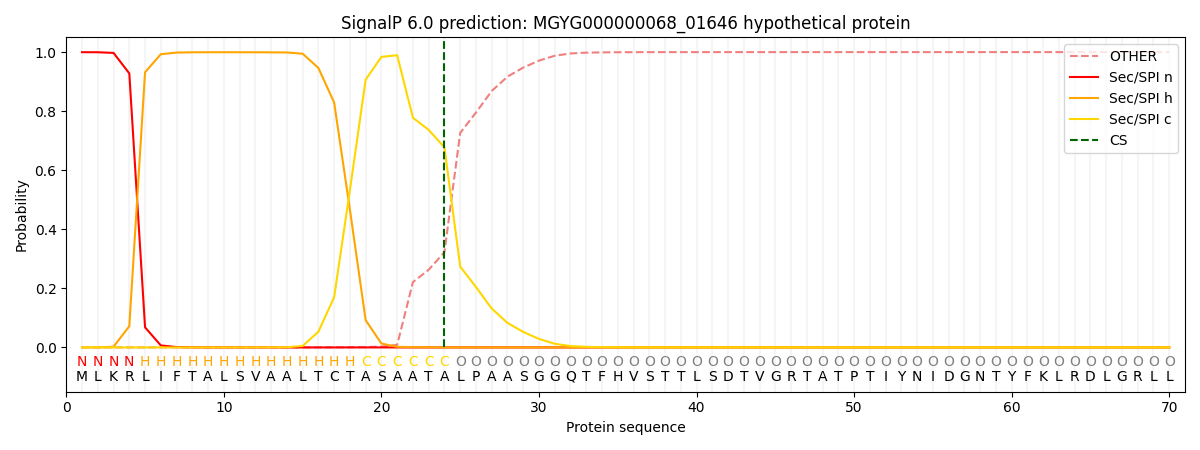
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000495 | 0.998154 | 0.000679 | 0.000268 | 0.000213 | 0.000179 |
