You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000071_00738
You are here: Home > Sequence: MGYG000000071_00738
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudoflavonifractor capillosus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Pseudoflavonifractor; Pseudoflavonifractor capillosus | |||||||||||
| CAZyme ID | MGYG000000071_00738 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 165724; End: 167031 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 143 | 369 | 8.8e-53 | 0.9629629629629629 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00933 | Glyco_hydro_3 | 4.63e-87 | 82 | 407 | 1 | 316 | Glycosyl hydrolase family 3 N terminal domain. |
| COG1472 | BglX | 5.04e-86 | 81 | 432 | 1 | 335 | Periplasmic beta-glucosidase and related glycosidases [Carbohydrate transport and metabolism]. |
| PRK05337 | PRK05337 | 4.23e-59 | 113 | 434 | 26 | 333 | beta-hexosaminidase; Provisional |
| PRK15098 | PRK15098 | 3.85e-22 | 59 | 417 | 17 | 362 | beta-glucosidase BglX. |
| TIGR00601 | rad23 | 0.004 | 7 | 83 | 70 | 150 | UV excision repair protein Rad23. All proteins in this family for which functions are known are components of a multiprotein complex used for targeting nucleotide excision repair to specific parts of the genome. In humans, Rad23 complexes with the XPC protein. This family is based on the phylogenomic analysis of JA Eisen (1999, Ph.D. Thesis, Stanford University). [DNA metabolism, DNA replication, recombination, and repair] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALP95074.1 | 6.61e-148 | 77 | 435 | 61 | 424 |
| QBB66908.1 | 6.61e-148 | 77 | 435 | 61 | 424 |
| QQR04853.1 | 8.64e-145 | 74 | 433 | 55 | 421 |
| ANU42253.1 | 8.64e-145 | 74 | 433 | 55 | 421 |
| QIA31937.1 | 1.40e-144 | 74 | 433 | 55 | 425 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6K5J_A | 2.68e-91 | 80 | 409 | 10 | 338 | Structureof a glycoside hydrolase family 3 beta-N-acetylglucosaminidase from Paenibacillus sp. str. FPU-7 [Paenibacillaceae] |
| 3BMX_A | 3.86e-67 | 81 | 413 | 42 | 398 | Beta-N-hexosaminidase(YbbD) from Bacillus subtilis [Bacillus subtilis],3BMX_B Beta-N-hexosaminidase (YbbD) from Bacillus subtilis [Bacillus subtilis],3NVD_A Structure of YBBD in complex with pugnac [Bacillus subtilis],3NVD_B Structure of YBBD in complex with pugnac [Bacillus subtilis] |
| 3LK6_A | 1.25e-66 | 81 | 413 | 16 | 372 | ChainA, Lipoprotein ybbD [Bacillus subtilis],3LK6_B Chain B, Lipoprotein ybbD [Bacillus subtilis],3LK6_C Chain C, Lipoprotein ybbD [Bacillus subtilis],3LK6_D Chain D, Lipoprotein ybbD [Bacillus subtilis] |
| 4GYJ_A | 2.28e-66 | 81 | 413 | 46 | 402 | Crystalstructure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYJ_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1) [Bacillus subtilis subsp. subtilis str. 168],4GYK_A Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168],4GYK_B Crystal structure of mutant (D318N) bacillus subtilis family 3 glycoside hydrolase (nagz) in complex with glcnac-murnac (space group P1211) [Bacillus subtilis subsp. subtilis str. 168] |
| 4ZM6_A | 1.17e-63 | 83 | 432 | 9 | 361 | Aunique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432],4ZM6_B A unique GCN5-related glucosamine N-acetyltransferase region exist in the fungal multi-domain GH3 beta-N-acetylglucosaminidase [Rhizomucor miehei CAU432] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P40406 | 2.11e-66 | 81 | 413 | 42 | 398 | Beta-hexosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=nagZ PE=1 SV=1 |
| Q606N2 | 9.37e-47 | 114 | 389 | 23 | 292 | Beta-hexosaminidase OS=Methylococcus capsulatus (strain ATCC 33009 / NCIMB 11132 / Bath) OX=243233 GN=nagZ PE=3 SV=1 |
| P48823 | 2.75e-46 | 104 | 431 | 55 | 410 | Beta-hexosaminidase A OS=Pseudoalteromonas piscicida OX=43662 GN=cht60 PE=1 SV=1 |
| Q3SKU2 | 4.86e-45 | 97 | 397 | 12 | 311 | Beta-hexosaminidase OS=Thiobacillus denitrificans (strain ATCC 25259) OX=292415 GN=nagZ PE=3 SV=1 |
| L7N6B0 | 1.82e-44 | 65 | 408 | 40 | 380 | Beta-hexosaminidase LpqI OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=lpqI PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
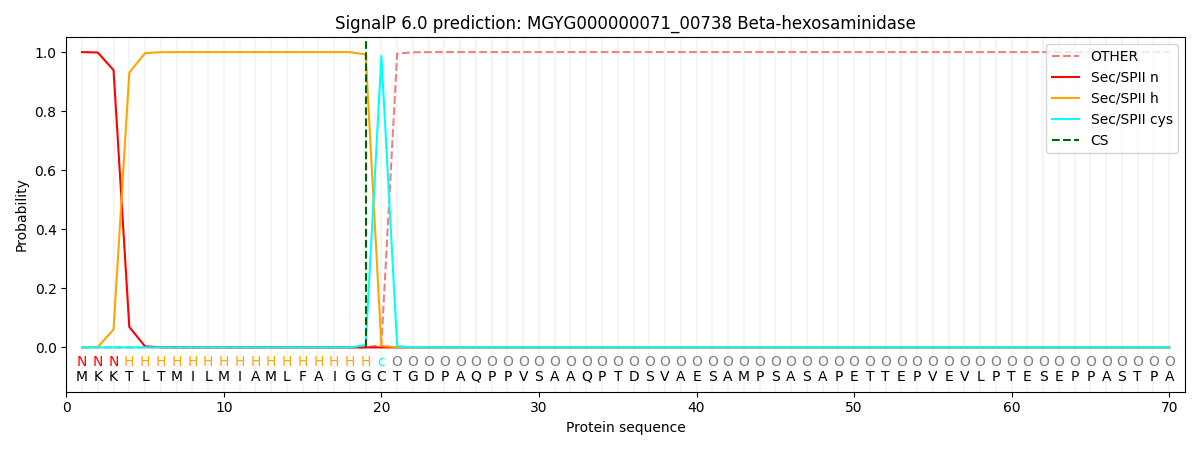
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000064 | 0.000000 | 0.000000 | 0.000000 |
