You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000071_00763
You are here: Home > Sequence: MGYG000000071_00763
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pseudoflavonifractor capillosus | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; Pseudoflavonifractor; Pseudoflavonifractor capillosus | |||||||||||
| CAZyme ID | MGYG000000071_00763 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 199631; End: 203236 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 57 | 845 | 1.5e-73 | 0.8009708737864077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 3.08e-12 | 234 | 761 | 349 | 837 | alpha-L-rhamnosidase. |
| pfam00395 | SLH | 5.11e-08 | 1072 | 1113 | 1 | 42 | S-layer homology domain. |
| NF033190 | inl_like_NEAT_1 | 1.73e-04 | 1050 | 1144 | 621 | 716 | NEAT domain-containing leucine-rich repeat protein. Members of this family have an N-terminal NEAT (near transporter) domain often associated with iron transport, followed by a leucine-rich repeat region with significant sequence similarity to the internalins of Listeria monocytogenes. However, since Bacillus cereus (from which this protein was described, in PMID:16978259) is not considered an intracellular pathogen, and the function may be iron transport rather than internalization, applying the name "internalin" to this family probably would be misleading. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH71057.1 | 4.84e-274 | 46 | 1067 | 77 | 1134 |
| AHF25191.1 | 1.56e-256 | 58 | 1059 | 65 | 1147 |
| QGH70002.1 | 2.51e-134 | 48 | 1067 | 84 | 1118 |
| SDS67158.1 | 2.77e-130 | 53 | 1030 | 5 | 943 |
| SDG73971.1 | 8.58e-130 | 53 | 1030 | 5 | 936 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 1.90e-10 | 71 | 681 | 46 | 601 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
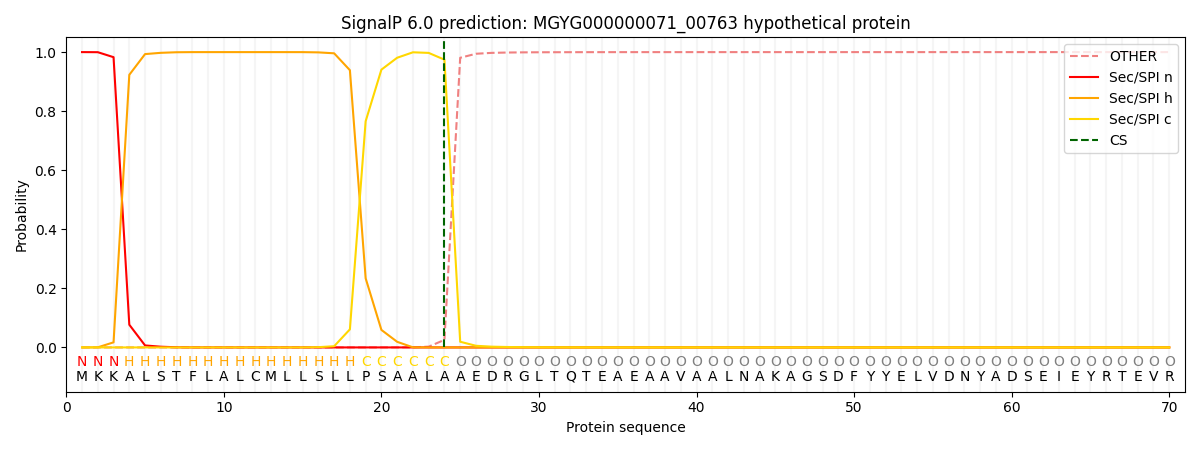
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000261 | 0.998916 | 0.000227 | 0.000204 | 0.000188 | 0.000176 |
