You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000072_00762
You are here: Home > Sequence: MGYG000000072_00762
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
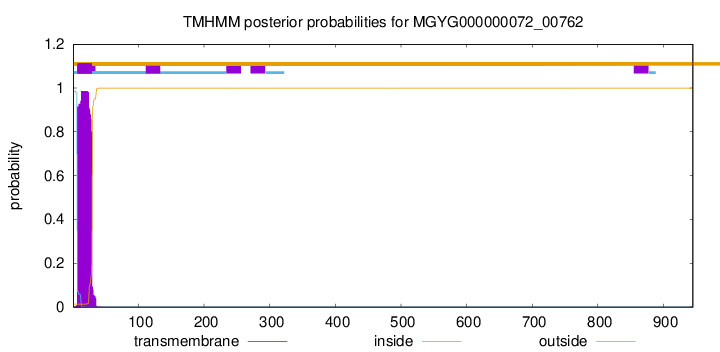
TMHMM annotations
Basic Information help
| Species | UBA1394 sp900066845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1394; UBA1394 sp900066845 | |||||||||||
| CAZyme ID | MGYG000000072_00762 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 145896; End: 148733 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 228 | 420 | 1.2e-30 | 0.8316831683168316 |
| CBM13 | 550 | 695 | 1.5e-22 | 0.7021276595744681 |
| CBM13 | 704 | 863 | 1e-16 | 0.776595744680851 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 2.06e-23 | 158 | 496 | 20 | 340 | Pectate lyase [Carbohydrate transport and metabolism]. |
| pfam14200 | RicinB_lectin_2 | 5.77e-18 | 635 | 733 | 1 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 6.68e-18 | 589 | 678 | 5 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| pfam14200 | RicinB_lectin_2 | 1.68e-17 | 548 | 629 | 12 | 89 | Ricin-type beta-trefoil lectin domain-like. |
| cd14256 | Dockerin_I | 7.96e-16 | 887 | 942 | 1 | 56 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CDM70399.1 | 8.96e-197 | 36 | 693 | 35 | 696 |
| AUO18238.1 | 2.22e-188 | 36 | 693 | 39 | 707 |
| CBL17651.1 | 3.33e-142 | 30 | 723 | 33 | 722 |
| CBL16867.1 | 3.18e-127 | 69 | 921 | 77 | 891 |
| ASR46637.1 | 6.59e-94 | 30 | 517 | 35 | 537 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1VBL_A | 1.80e-10 | 265 | 390 | 156 | 301 | Structureof the thermostable pectate lyase PL 47 [Bacillus sp. TS-47] |
| 1PCL_A | 7.10e-09 | 182 | 449 | 14 | 316 | ChainA, PECTATE LYASE E [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P94449 | 2.03e-19 | 168 | 496 | 35 | 335 | Pectin lyase OS=Bacillus subtilis OX=1423 GN=pelB PE=1 SV=1 |
| Q2TZY0 | 1.79e-17 | 182 | 389 | 43 | 234 | Probable pectate lyase B OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyB PE=3 SV=1 |
| B8NBC2 | 1.79e-17 | 182 | 389 | 43 | 234 | Probable pectate lyase B OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyB PE=3 SV=1 |
| Q5AVN4 | 2.17e-14 | 185 | 451 | 51 | 303 | Pectate lyase A OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyA PE=1 SV=1 |
| Q00645 | 2.87e-14 | 182 | 389 | 43 | 234 | Pectate lyase plyB OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
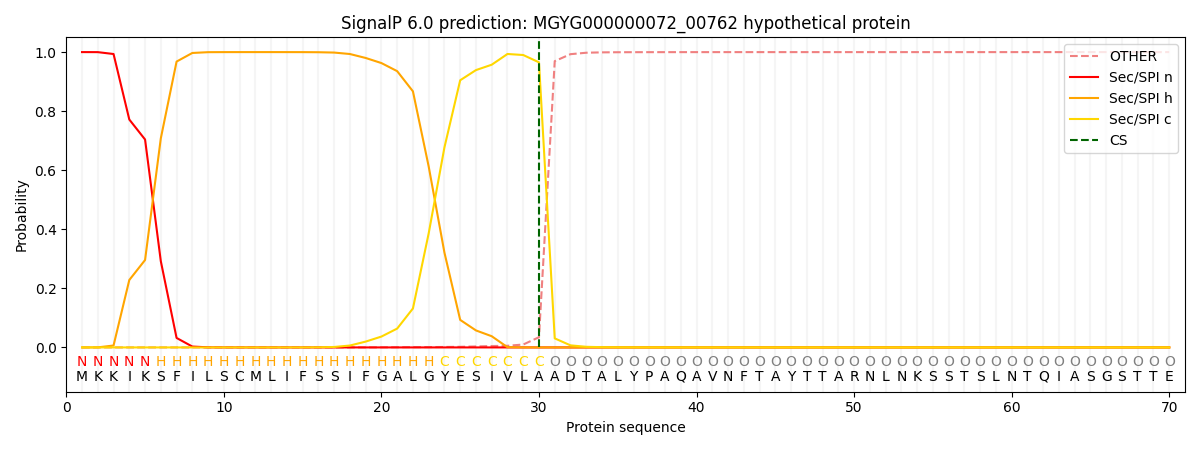
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000367 | 0.998945 | 0.000200 | 0.000160 | 0.000152 | 0.000145 |