You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000072_01154
You are here: Home > Sequence: MGYG000000072_01154
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
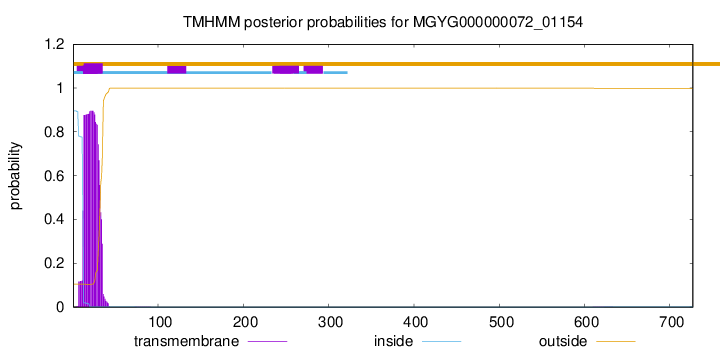
TMHMM annotations
Basic Information help
| Species | UBA1394 sp900066845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1394; UBA1394 sp900066845 | |||||||||||
| CAZyme ID | MGYG000000072_01154 | |||||||||||
| CAZy Family | CBM79 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 131222; End: 133405 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM79 | 412 | 522 | 3.2e-21 | 0.9545454545454546 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18522 | DUF5620 | 7.33e-22 | 234 | 382 | 1 | 119 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| pfam18522 | DUF5620 | 2.36e-19 | 412 | 527 | 1 | 116 | Domain of unknown function (DUF5620). This is a domain of unknown function predicted to be a carbohydrate binding module. |
| cd14256 | Dockerin_I | 1.16e-06 | 646 | 705 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam02368 | Big_2 | 1.30e-06 | 540 | 600 | 4 | 71 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| COG5492 | YjdB | 0.002 | 541 | 600 | 186 | 253 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL17555.1 | 6.30e-108 | 8 | 536 | 7 | 511 |
| EWM54821.1 | 2.27e-71 | 3 | 530 | 2 | 543 |
| CCZ83178.1 | 2.31e-64 | 89 | 721 | 161 | 875 |
| CDE32105.1 | 2.88e-25 | 238 | 525 | 90 | 336 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
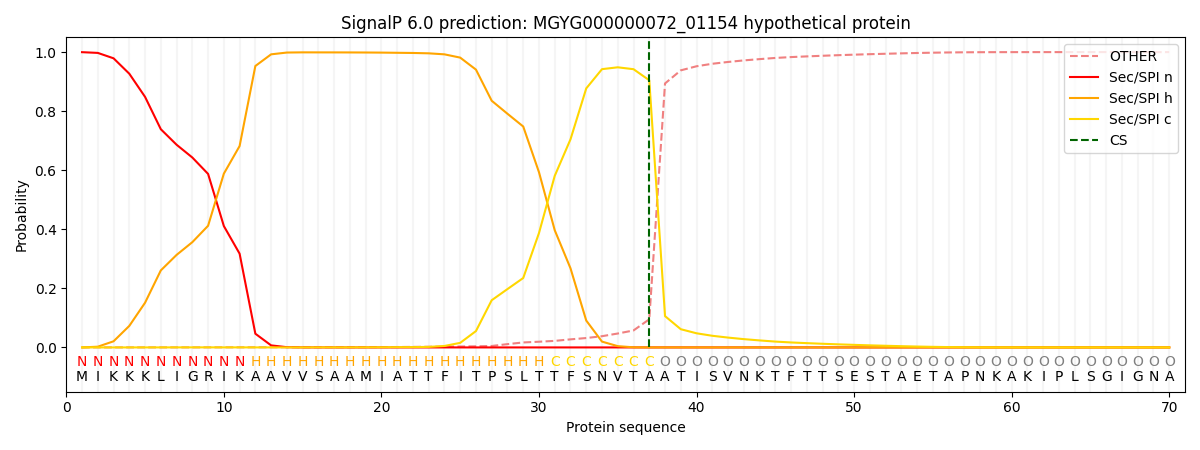
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000667 | 0.998503 | 0.000229 | 0.000211 | 0.000173 | 0.000158 |