You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000072_01857
You are here: Home > Sequence: MGYG000000072_01857
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
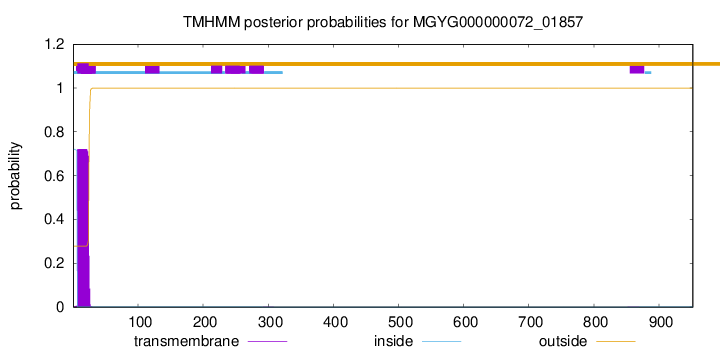
TMHMM annotations
Basic Information help
| Species | UBA1394 sp900066845 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; UBA1394; UBA1394 sp900066845 | |||||||||||
| CAZyme ID | MGYG000000072_01857 | |||||||||||
| CAZy Family | GH30 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30445; End: 33303 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH30 | 34 | 386 | 9e-130 | 0.9883040935672515 |
| CE1 | 715 | 947 | 1e-38 | 0.9691629955947136 |
| CBM22 | 448 | 573 | 1.2e-28 | 0.9465648854961832 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5520 | XynC | 2.69e-117 | 2 | 432 | 4 | 430 | O-Glycosyl hydrolase [Cell wall/membrane/envelope biogenesis]. |
| COG2382 | Fes | 6.64e-24 | 658 | 952 | 22 | 299 | Enterochelin esterase or related enzyme [Inorganic ion transport and metabolism]. |
| pfam00756 | Esterase | 2.12e-18 | 714 | 941 | 1 | 239 | Putative esterase. This family contains Esterase D. However it is not clear if all members of the family have the same function. This family is related to the pfam00135 family. |
| pfam02018 | CBM_4_9 | 2.51e-18 | 449 | 577 | 8 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
| COG2819 | YbbA | 4.00e-14 | 714 | 862 | 16 | 171 | Predicted hydrolase of the alpha/beta superfamily [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL17903.1 | 0.0 | 4 | 951 | 7 | 955 |
| ADU21034.1 | 5.67e-226 | 28 | 598 | 4 | 575 |
| BAB39494.1 | 1.51e-225 | 28 | 598 | 33 | 604 |
| CBL17231.1 | 1.29e-176 | 437 | 951 | 560 | 1074 |
| BAB39495.1 | 6.26e-160 | 154 | 598 | 1 | 447 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3GTN_A | 3.92e-136 | 32 | 430 | 2 | 389 | CrystalStructure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3GTN_B Crystal Structure of XynC from Bacillus subtilis 168 [Bacillus subtilis],3KL0_A Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_B Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_C Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL0_D Crystal structure of the glucuronoxylan xylanohydrolase XynC from Bacillus subtilis [Bacillus subtilis subsp. subtilis str. 168],3KL3_A Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_B Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_C Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL3_D Crystal structure of Ligand bound XynC [Bacillus subtilis subsp. subtilis str. 168],3KL5_A Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_B Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_C Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis],3KL5_D Structure Analysis of a Xylanase From Glycosyl Hydrolase Family Thirty: Carbohydrate Ligand Complexes Reveal this Family of Enzymes Unique Mechanism of Substrate Specificity and Recognition [Bacillus subtilis] |
| 4QAW_A | 1.13e-128 | 32 | 433 | 1 | 391 | Structureof modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_B Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_C Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_D Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_E Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_F Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_G Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4QAW_H Structure of modular Xyn30D from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 4CKQ_A | 1.01e-118 | 33 | 429 | 22 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQ9_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQB_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQC_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQD_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],4UQE_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 4UQA_A | 2.00e-118 | 33 | 429 | 22 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus] |
| 5A6L_A | 7.75e-118 | 33 | 429 | 22 | 409 | ChainA, Carbohydrate Binding Family 6 [Acetivibrio thermocellus],5A6M_A Chain A, Carbohydrate Binding Family 6 [Acetivibrio thermocellus ATCC 27405] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q45070 | 1.12e-135 | 31 | 430 | 32 | 420 | Glucuronoxylanase XynC OS=Bacillus subtilis (strain 168) OX=224308 GN=xynC PE=1 SV=1 |
| Q6YK37 | 1.37e-129 | 30 | 430 | 32 | 421 | Glucuronoxylanase XynC OS=Bacillus subtilis OX=1423 GN=xynC PE=3 SV=2 |
| P51584 | 1.96e-62 | 439 | 855 | 562 | 973 | Endo-1,4-beta-xylanase Y OS=Acetivibrio thermocellus OX=1515 GN=xynY PE=1 SV=1 |
| P10478 | 4.30e-31 | 653 | 951 | 13 | 279 | Endo-1,4-beta-xylanase Z OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xynZ PE=1 SV=3 |
| Q53317 | 1.11e-29 | 435 | 667 | 251 | 512 | Xylanase/beta-glucanase OS=Ruminococcus flavefaciens OX=1265 GN=xynD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
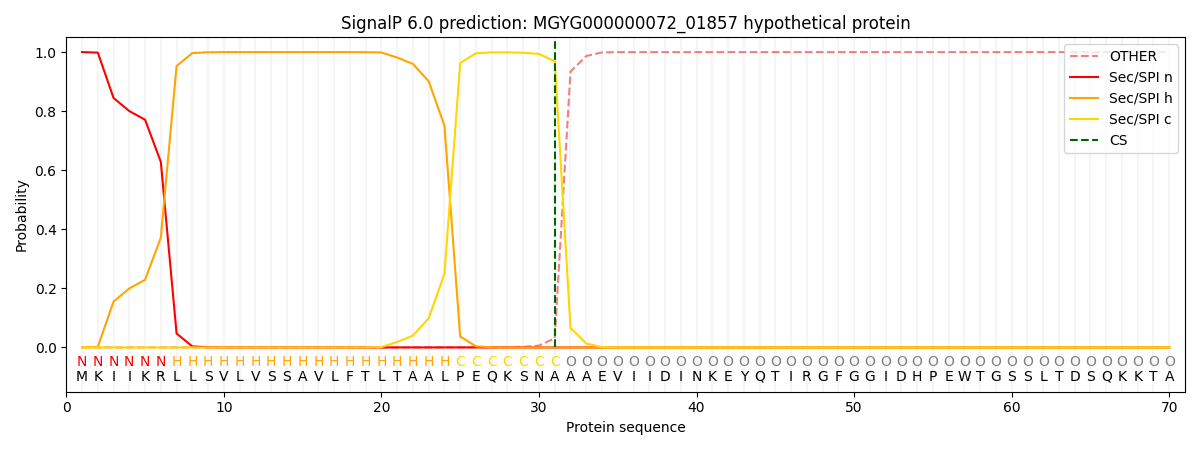
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000251 | 0.998966 | 0.000306 | 0.000167 | 0.000160 | 0.000139 |