You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000073_01033
You are here: Home > Sequence: MGYG000000073_01033
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | GCA-900066905 sp900066905 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; GCA-900066905; GCA-900066905; GCA-900066905 sp900066905 | |||||||||||
| CAZyme ID | MGYG000000073_01033 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 376781; End: 380749 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 856 | 1084 | 1.9e-54 | 0.9013452914798207 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 7.53e-23 | 1098 | 1316 | 4 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 2.21e-17 | 1082 | 1320 | 490 | 695 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG4346 | COG4346 | 3.88e-16 | 877 | 1134 | 132 | 330 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
| COG5542 | COG5542 | 7.56e-16 | 303 | 595 | 92 | 371 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| COG1928 | PMT1 | 2.13e-15 | 840 | 1082 | 50 | 264 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CQR52353.1 | 4.07e-242 | 17 | 1319 | 17 | 1274 |
| AIQ27397.1 | 1.94e-240 | 27 | 1319 | 28 | 1278 |
| AIQ39188.1 | 1.07e-239 | 27 | 1319 | 28 | 1278 |
| AIQ56104.1 | 8.28e-239 | 27 | 1319 | 28 | 1278 |
| QUL55595.1 | 4.43e-238 | 27 | 1319 | 28 | 1277 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P9WN04 | 7.55e-20 | 858 | 1318 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 7.55e-20 | 858 | 1318 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| Q59X23 | 1.87e-06 | 1081 | 1252 | 534 | 670 | Dolichyl-phosphate-mannose--protein mannosyltransferase 4 OS=Candida albicans (strain SC5314 / ATCC MYA-2876) OX=237561 GN=PMT4 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000210 | 0.999097 | 0.000182 | 0.000168 | 0.000155 | 0.000147 |
TMHMM Annotations download full data without filtering help
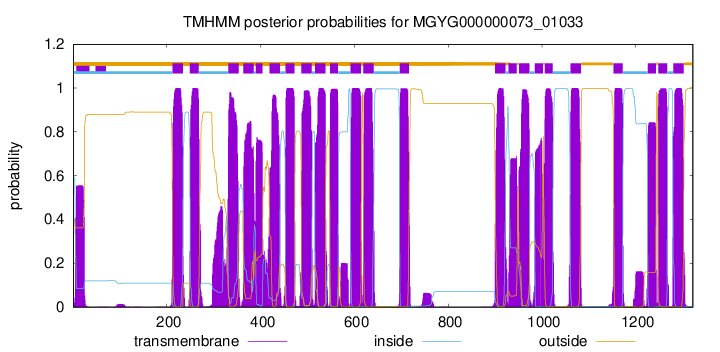
| start | end |
|---|---|
| 212 | 234 |
| 249 | 268 |
| 331 | 353 |
| 363 | 385 |
| 390 | 404 |
| 419 | 441 |
| 453 | 472 |
| 487 | 509 |
| 516 | 538 |
| 548 | 565 |
| 592 | 614 |
| 619 | 641 |
| 697 | 716 |
| 900 | 922 |
| 929 | 946 |
| 951 | 973 |
| 985 | 1002 |
| 1006 | 1023 |
| 1061 | 1083 |
| 1153 | 1172 |
| 1226 | 1243 |
| 1248 | 1267 |
| 1280 | 1302 |
