You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000074_01404
You are here: Home > Sequence: MGYG000000074_01404
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes onderdonkii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes onderdonkii | |||||||||||
| CAZyme ID | MGYG000000074_01404 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 162577; End: 163782 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 36 | 368 | 4.6e-54 | 0.9050279329608939 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 2.73e-20 | 140 | 300 | 85 | 257 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG1331 | YyaL | 1.71e-07 | 143 | 358 | 274 | 512 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| COG1331 | YyaL | 2.91e-07 | 130 | 292 | 401 | 565 | Uncharacterized conserved protein YyaL, SSP411 family, contains thoiredoxin and six-hairpin glycosidase-like domains [General function prediction only]. |
| COG4833 | COG4833 | 4.20e-06 | 139 | 305 | 90 | 252 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| cd04792 | LanM-like | 5.95e-05 | 141 | 339 | 493 | 676 | Cyclases involved in the biosynthesis of class II lantibiotics, and similar proteins. LanM-like proteins. LanM is a bifunctional enzyme, involved in the synthesis of class II lantibiotics. It is responsible for both the dehydration and the cyclization of the precursor-peptide during lantibiotic synthesis. The C-terminal domain shows similarity to LanC, the cyclase component of the lan operon, but the N terminus seems to be unrelated to the dehydratase, LanB. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL07983.1 | 2.00e-310 | 1 | 401 | 1 | 401 |
| BBL10774.1 | 2.00e-310 | 1 | 401 | 1 | 401 |
| BBK99968.1 | 2.15e-305 | 1 | 401 | 1 | 401 |
| QUT25297.1 | 1.06e-170 | 35 | 394 | 34 | 392 |
| QDM09503.1 | 1.10e-170 | 35 | 394 | 34 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MU9_A | 1.19e-65 | 33 | 384 | 6 | 359 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 4C1S_A | 1.14e-30 | 67 | 393 | 30 | 373 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6U4Z_A | 8.17e-26 | 57 | 337 | 155 | 441 | CrystalStructure of a family 76 glycoside hydrolase from a bovine Bacteroides thetaiotaomicron strain [Bacteroides thetaiotaomicron] |
| 6SHD_A | 1.36e-16 | 140 | 291 | 133 | 286 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 1.09e-15 | 107 | 291 | 96 | 287 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
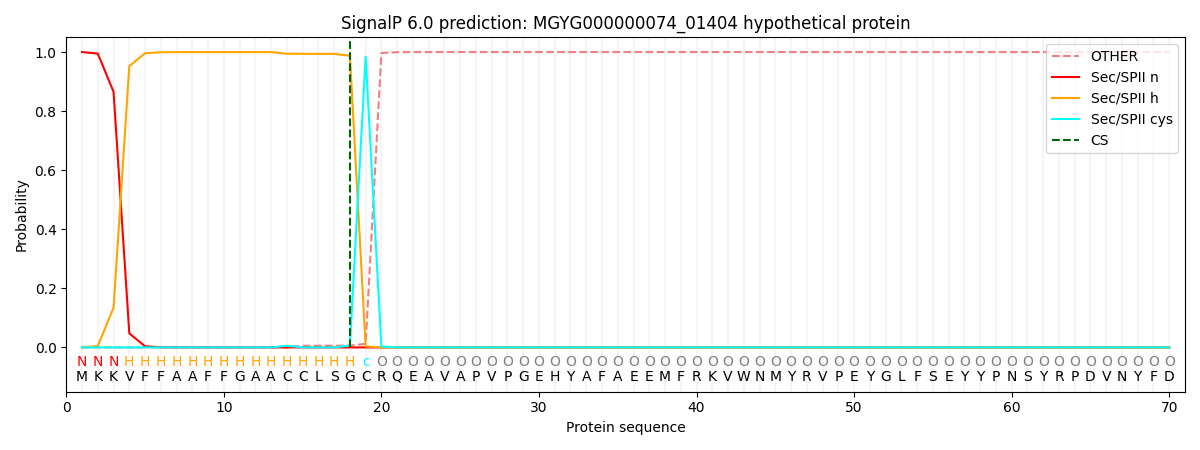
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000009 | 1.000044 | 0.000000 | 0.000000 | 0.000000 |
