You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000074_02136
You are here: Home > Sequence: MGYG000000074_02136
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes onderdonkii | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes onderdonkii | |||||||||||
| CAZyme ID | MGYG000000074_02136 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 322827; End: 324236 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 67 | 320 | 4.1e-21 | 0.6690909090909091 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 0.002 | 78 | 227 | 39 | 181 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL00987.1 | 0.0 | 1 | 469 | 1 | 469 |
| BBL08912.1 | 0.0 | 1 | 469 | 1 | 469 |
| BBL11704.1 | 0.0 | 1 | 469 | 1 | 469 |
| ASV75818.1 | 3.82e-141 | 33 | 469 | 32 | 469 |
| AXE20299.1 | 3.75e-132 | 11 | 468 | 10 | 474 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NIV9 | 2.03e-09 | 65 | 239 | 151 | 335 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
| Q2U2I3 | 2.69e-09 | 65 | 239 | 151 | 335 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manF PE=3 SV=2 |
| A1C8U0 | 1.32e-07 | 105 | 239 | 181 | 310 | Mannan endo-1,4-beta-mannosidase F OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=manF PE=3 SV=1 |
| Q5AVP1 | 1.46e-06 | 1 | 298 | 1 | 301 | Mannan endo-1,4-beta-mannosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manD PE=3 SV=1 |
| B0Y9E7 | 3.88e-06 | 77 | 239 | 168 | 313 | Probable mannan endo-1,4-beta-mannosidase F OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=manF PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
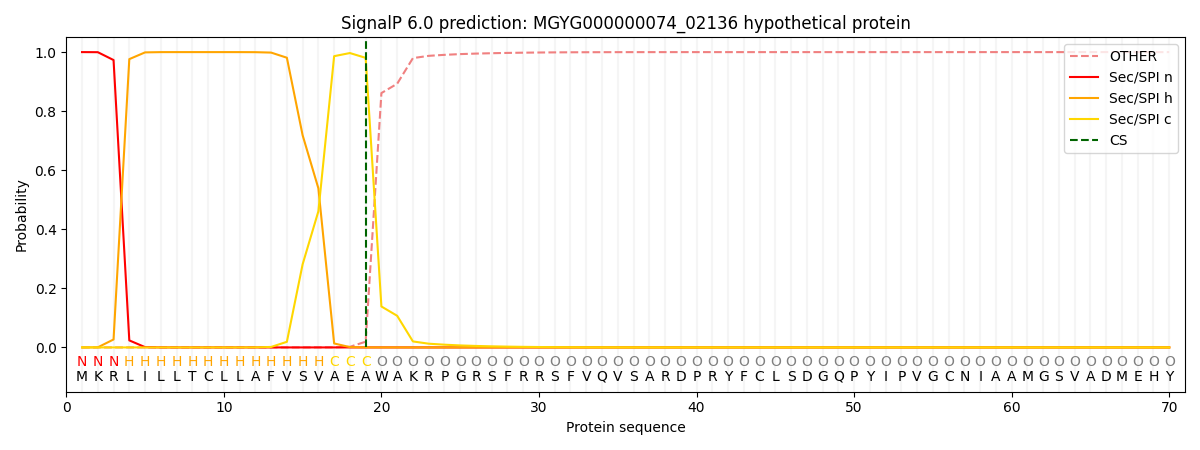
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000291 | 0.999057 | 0.000184 | 0.000159 | 0.000145 | 0.000137 |
