You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000079_00172
You are here: Home > Sequence: MGYG000000079_00172
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exiguobacterium sp902362975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Exiguobacterales; Exiguobacteraceae; Exiguobacterium; Exiguobacterium sp902362975 | |||||||||||
| CAZyme ID | MGYG000000079_00172 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 157423; End: 160395 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 8.03e-74 | 765 | 990 | 45 | 245 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| smart00047 | LYZ2 | 2.20e-28 | 811 | 980 | 1 | 146 | Lysozyme subfamily 2. Eubacterial enzymes distantly related to eukaryotic lysozymes. |
| pfam01832 | Glucosaminidase | 1.50e-07 | 823 | 925 | 1 | 78 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
| COG3103 | YgiM | 2.99e-05 | 14 | 168 | 7 | 151 | Uncharacterized conserved protein YgiM, contains N-terminal SH3 domain, DUF1202 family [General function prediction only]. |
| pfam08239 | SH3_3 | 3.77e-05 | 114 | 165 | 2 | 52 | Bacterial SH3 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QPI67182.1 | 0.0 | 1 | 990 | 1 | 989 |
| ACQ71242.1 | 0.0 | 1 | 990 | 1 | 990 |
| QUE86191.1 | 0.0 | 1 | 990 | 1 | 853 |
| QUP86639.1 | 7.61e-314 | 1 | 990 | 1 | 770 |
| AJD93634.1 | 1.59e-92 | 404 | 989 | 65 | 562 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FXO_A | 2.17e-46 | 744 | 990 | 21 | 244 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
| 6U0O_B | 3.78e-36 | 765 | 977 | 73 | 263 | ChainB, LYZ2 domain-containing protein [Staphylococcus aureus subsp. aureus NCTC 8325] |
| 6FXP_A | 5.84e-36 | 765 | 977 | 43 | 233 | ChainA, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50],6FXP_B Chain B, Uncharacterized protein [Staphylococcus aureus subsp. aureus Mu50] |
| 5WQW_A | 7.55e-31 | 772 | 989 | 43 | 269 | X-raystructure of catalytic domain of autolysin from Clostridium perfringens [Clostridium perfringens str. 13] |
| 4PI7_A | 1.61e-30 | 794 | 965 | 55 | 203 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 2.95e-63 | 707 | 990 | 627 | 880 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q6GAG0 | 4.08e-41 | 744 | 990 | 1027 | 1250 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
| Q8NX96 | 4.11e-41 | 744 | 990 | 1033 | 1256 | Bifunctional autolysin OS=Staphylococcus aureus (strain MW2) OX=196620 GN=atl PE=3 SV=1 |
| Q99V41 | 5.38e-41 | 744 | 990 | 1025 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain N315) OX=158879 GN=atl PE=1 SV=1 |
| Q931U5 | 5.38e-41 | 744 | 990 | 1025 | 1248 | Bifunctional autolysin OS=Staphylococcus aureus (strain Mu50 / ATCC 700699) OX=158878 GN=atl PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
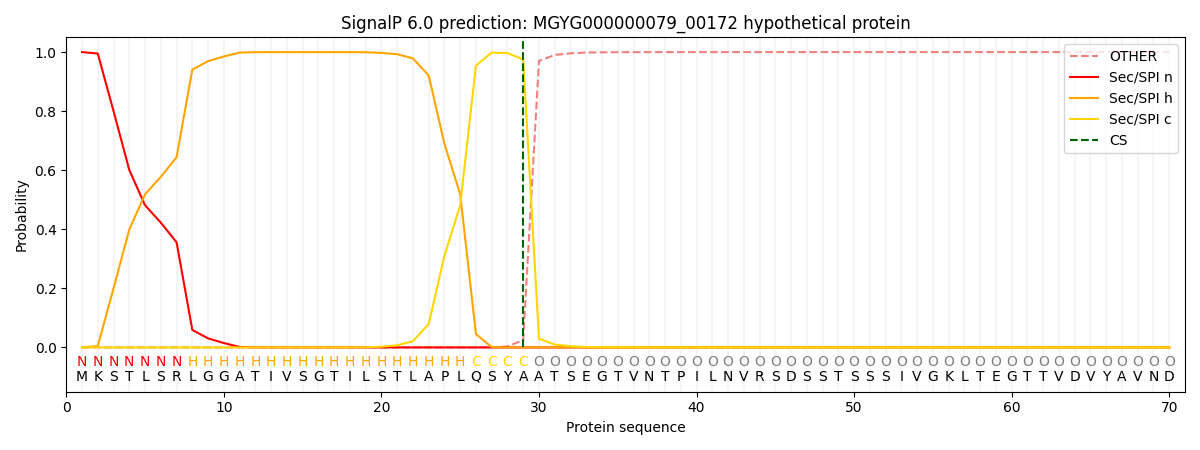
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000335 | 0.998863 | 0.000193 | 0.000247 | 0.000188 | 0.000159 |
