You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000079_02545
You are here: Home > Sequence: MGYG000000079_02545
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Exiguobacterium sp902362975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Exiguobacterales; Exiguobacteraceae; Exiguobacterium; Exiguobacterium sp902362975 | |||||||||||
| CAZyme ID | MGYG000000079_02545 | |||||||||||
| CAZy Family | GH166 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 896; End: 1753 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH166 | 61 | 280 | 1.5e-41 | 0.9955357142857143 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03537 | Glyco_hydro_114 | 2.34e-08 | 60 | 160 | 20 | 109 | Glycoside-hydrolase family GH114. This family is recognized as a glycosyl-hydrolase family, number 114. It is endo-alpha-1,4-polygalactosaminidase, a rare enzyme. It is proposed to be TIM-barrel, the most common structure amongst the catalytic domains of glycosyl-hydrolases. |
| COG2342 | COG2342 | 4.49e-07 | 20 | 160 | 3 | 148 | Endo alpha-1,4 polygalactosaminidase, GH114 family (was erroneously annotated as Cys-tRNA synthetase) [Carbohydrate transport and metabolism]. |
| pfam14885 | GHL15 | 1.99e-04 | 123 | 232 | 61 | 174 | Hypothetical glycosyl hydrolase family 15. GHL15 is a family of hypothetical glycoside hydrolases. |
| COG3868 | COG3868 | 3.16e-04 | 37 | 178 | 56 | 192 | Predicted glycosyl hydrolase, GH114 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QPI68214.1 | 2.22e-155 | 1 | 285 | 1 | 285 |
| QUP87669.1 | 2.22e-155 | 1 | 285 | 1 | 285 |
| ACQ70142.1 | 1.28e-154 | 1 | 285 | 1 | 285 |
| ASI36427.1 | 3.16e-134 | 1 | 285 | 1 | 285 |
| AHA30789.1 | 1.73e-131 | 1 | 285 | 1 | 285 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
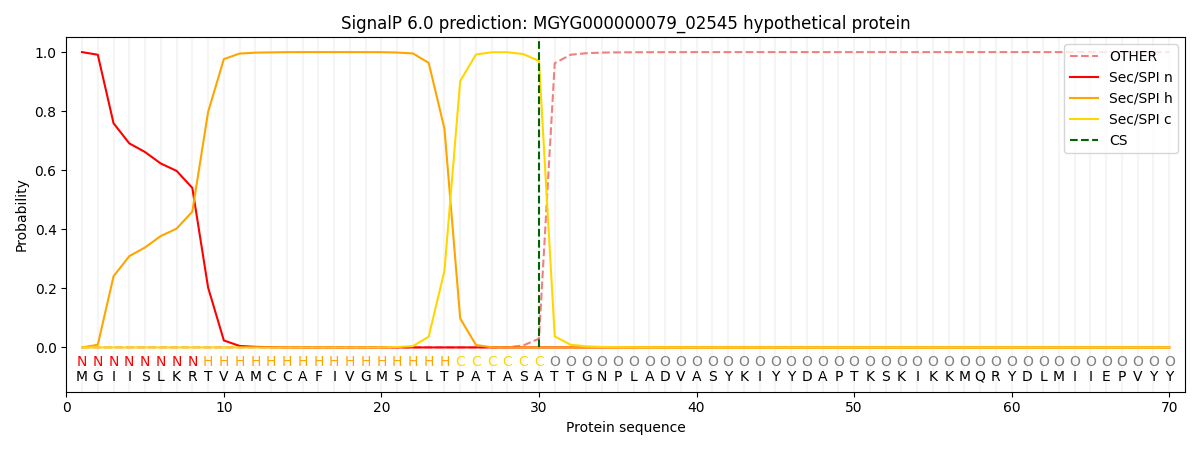
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000480 | 0.998633 | 0.000263 | 0.000233 | 0.000201 | 0.000183 |
