You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000089_00635
You are here: Home > Sequence: MGYG000000089_00635
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-353 sp900066885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; CAG-353; CAG-353 sp900066885 | |||||||||||
| CAZyme ID | MGYG000000089_00635 | |||||||||||
| CAZy Family | GH74 | |||||||||||
| CAZyme Description | Xyloglucanase Xgh74A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 600975; End: 603230 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH74 | 237 | 397 | 4.3e-34 | 0.6824034334763949 |
| GH74 | 120 | 230 | 1.2e-17 | 0.463519313304721 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam15902 | Sortilin-Vps10 | 4.60e-07 | 153 | 274 | 5 | 112 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| pfam15902 | Sortilin-Vps10 | 6.84e-06 | 517 | 614 | 6 | 110 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| pfam15902 | Sortilin-Vps10 | 2.60e-05 | 562 | 725 | 4 | 179 | Sortilin, neurotensin receptor 3,. Sortilin, also known in mammals as neurotensin receptor-3, is the archetypical member of a Vps10-domain (Vps10-D) that binds neurotrophic factors and neuropeptides. This domain constitutes the entire luminal part of Sortilin and is activated in the trans-Golgi network by enzymatic propeptide cleavage. The structure of the domain has been determined as a ten-bladed propeller, with up to 9 BNR or beta-hairpin turns in it. The mature receptor binds various ligands, including its own propeptide (Sort-pro), neurotensin, the pro-forms of nerve growth factor-beta (NGF)6 and brain-derived neurotrophic factor (BDNF)7, lipoprotein lipase (LpL), apo lipoprotein AV14 and the receptor-associated protein (RAP)1. |
| COG4447 | COG4447 | 6.36e-05 | 517 | 660 | 153 | 298 | Uncharacterized protein related to plant photosystem II stability/assembly factor [General function prediction only]. |
| cd15482 | Sialidase_non-viral | 0.002 | 516 | 638 | 178 | 320 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL33465.1 | 5.02e-245 | 54 | 747 | 57 | 747 |
| CBK97166.1 | 3.66e-244 | 61 | 747 | 61 | 744 |
| QJB39025.1 | 8.27e-176 | 65 | 747 | 65 | 743 |
| AHM63515.1 | 1.28e-175 | 60 | 745 | 50 | 731 |
| QJB32550.1 | 9.16e-175 | 65 | 747 | 65 | 743 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6P2L_A | 4.90e-153 | 62 | 743 | 7 | 681 | Crystalstructure of Niastella koreensis GH74 (NkGH74) enzyme [Niastella koreensis GR20-10] |
| 6P2M_A | 3.72e-118 | 54 | 745 | 3 | 690 | ChainA, Type 3a cellulose-binding domain protein [Caldicellulosiruptor lactoaceticus 6A] |
| 7KN8_A | 3.81e-110 | 61 | 747 | 10 | 713 | ChainA, Cellulase [Xanthomonas campestris pv. campestris str. ATCC 33913],7KN8_B Chain B, Cellulase [Xanthomonas campestris pv. campestris str. ATCC 33913] |
| 2CN2_A | 1.53e-99 | 69 | 745 | 20 | 728 | ChainA, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus],2CN2_B Chain B, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus],2CN2_C Chain C, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus],2CN2_D Chain D, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus] |
| 2CN3_A | 2.13e-99 | 69 | 745 | 20 | 728 | ChainA, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus],2CN3_B Chain B, BETA-1,4-XYLOGLUCAN HYDROLASE [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q70DK5 | 8.29e-99 | 69 | 745 | 47 | 755 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus OX=1515 GN=xghA PE=1 SV=1 |
| A3DFA0 | 3.13e-98 | 69 | 745 | 47 | 755 | Xyloglucanase Xgh74A OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=xghA PE=3 SV=1 |
| Q3MUH7 | 1.32e-97 | 70 | 747 | 48 | 765 | Xyloglucanase OS=Paenibacillus sp. OX=58172 GN=xeg74 PE=1 SV=1 |
| Q8J0D2 | 1.01e-88 | 61 | 751 | 27 | 788 | Oligoxyloglucan reducing end-specific cellobiohydrolase OS=Geotrichum sp. (strain M128) OX=203496 PE=1 SV=1 |
| A1DAU0 | 5.45e-83 | 61 | 747 | 27 | 797 | Probable oligoxyloglucan-reducing end-specific xyloglucanase OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=xgcA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
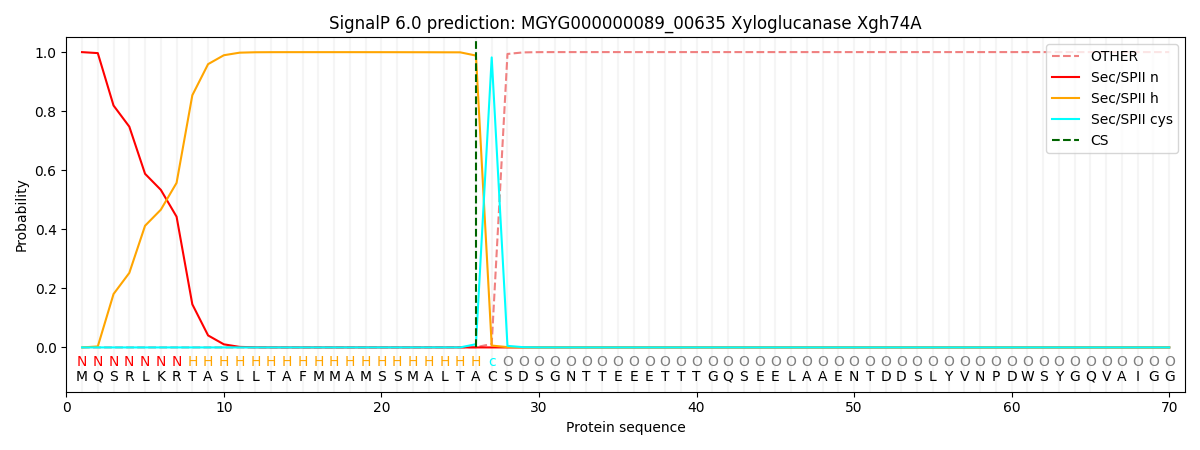
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000126 | 0.999907 | 0.000000 | 0.000000 | 0.000000 |
