You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000089_01943
You are here: Home > Sequence: MGYG000000089_01943
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-353 sp900066885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; CAG-353; CAG-353 sp900066885 | |||||||||||
| CAZyme ID | MGYG000000089_01943 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 420089; End: 421552 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 178 | 465 | 2.2e-89 | 0.9855072463768116 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.63e-46 | 182 | 434 | 19 | 244 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 5.55e-20 | 169 | 486 | 57 | 377 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd00063 | FN3 | 6.81e-07 | 41 | 128 | 2 | 93 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
| smart00060 | FN3 | 1.17e-04 | 42 | 113 | 3 | 82 | Fibronectin type 3 domain. One of three types of internal repeat within the plasma protein, fibronectin. The tenth fibronectin type III repeat contains a RGD cell recognition sequence in a flexible loop between 2 strands. Type III modules are present in both extracellular and intracellular proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO05195.1 | 1.36e-109 | 130 | 487 | 33 | 394 |
| VEU81114.1 | 2.54e-98 | 145 | 473 | 34 | 362 |
| CBK96866.1 | 4.21e-98 | 139 | 472 | 35 | 366 |
| QOS40601.1 | 5.65e-92 | 150 | 478 | 49 | 369 |
| AHF25528.1 | 1.83e-90 | 145 | 473 | 16 | 354 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6XRK_A | 1.47e-54 | 145 | 484 | 24 | 365 | GH5-4broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate],6XRK_B GH5-4 broad specificity endoglucanase from an uncultured bovine rumen ciliate [uncultured bovine rumen ciliate] |
| 6WQP_A | 1.88e-52 | 155 | 433 | 26 | 296 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 2JEP_A | 2.06e-49 | 139 | 465 | 27 | 362 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 5OYC_A | 2.11e-49 | 138 | 433 | 38 | 326 | GH5endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYC_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYD_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_A GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107],5OYE_B GH5 endo-xyloglucanase from Cellvibrio japonicus [Cellvibrio japonicus Ueda107] |
| 6MQ4_A | 1.51e-48 | 136 | 427 | 3 | 282 | ChainA, cellulase [Acetivibrio cellulolyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A7LXT7 | 3.79e-47 | 148 | 427 | 153 | 432 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
| O08342 | 6.75e-47 | 145 | 427 | 40 | 330 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P54937 | 3.13e-43 | 148 | 427 | 42 | 311 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P28623 | 5.78e-43 | 148 | 427 | 46 | 305 | Endoglucanase D OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engD PE=1 SV=2 |
| P28621 | 1.52e-42 | 134 | 461 | 31 | 344 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
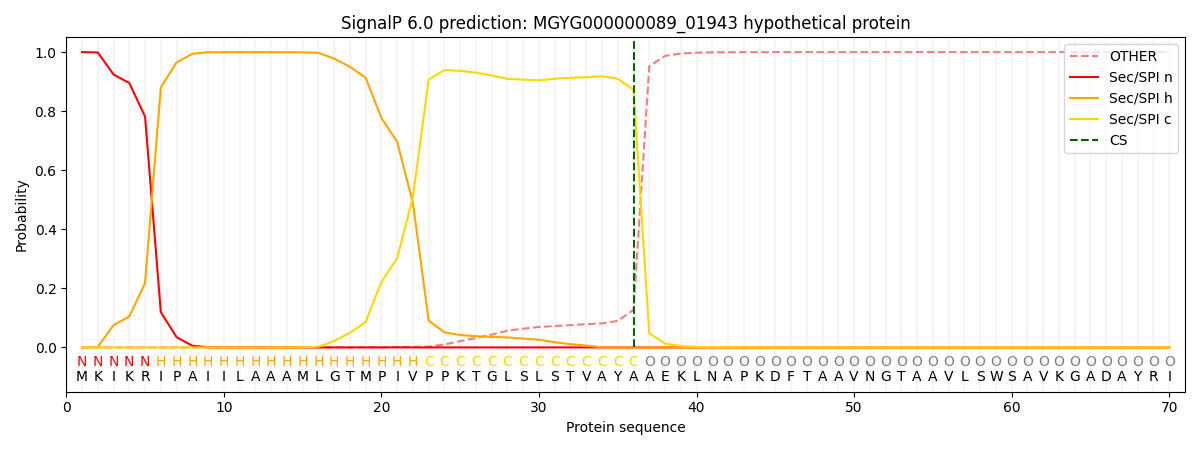
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000349 | 0.998940 | 0.000177 | 0.000207 | 0.000155 | 0.000140 |
