You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000091_00219
You are here: Home > Sequence: MGYG000000091_00219
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
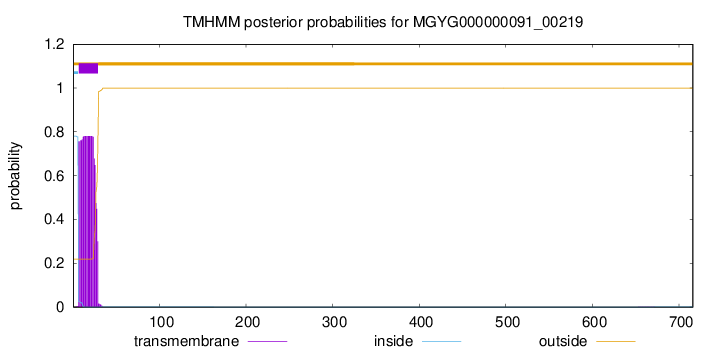
TMHMM annotations
Basic Information help
| Species | Achromobacter xylosoxidans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Achromobacter; Achromobacter xylosoxidans | |||||||||||
| CAZyme ID | MGYG000000091_00219 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | Non-hemolytic phospholipase C | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 254679; End: 256829 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR03396 | PC_PLC | 0.0 | 3 | 705 | 1 | 689 | phospholipase C, phosphocholine-specific, Pseudomonas-type. Members of this protein family are bacterial, phosphatidylcholine-hydrolyzing phospholipase C enzymes, with a characteristic domain architecture as found in hemolytyic (PlcH) and nonhemolytic (PlcN) secreted enzymes of Pseudomonas aeruginosa. PlcH hydrolyzes phosphatidylcholine to diacylglycerol and phosphocholine, but unlike PlcN can also hydrolyze sphingomyelin to ceramide ((N-acylsphingosine)) and phosphocholine. Members of this family share the twin-arginine signal sequence for Sec-independent transport across the plasma membrane. PlcH is secreted as a heterodimer with a small chaperone, PlcR, encoded immediately downstream. [Cellular processes, Pathogenesis] |
| COG3511 | PlcC | 1.33e-179 | 3 | 557 | 5 | 527 | Phospholipase C [Cell wall/membrane/envelope biogenesis]. |
| cd16014 | PLC | 6.94e-128 | 46 | 446 | 1 | 287 | non-hemolytic phospholipase C. Nonhemolytic Phospholipases C is produced by pathogenic bacterial. The toxic phospholipases C can interact with eukaryotic cell membranes and hydrolyze phosphatidylcholine and sphingomyelin, leading to cell lysis. |
| pfam04185 | Phosphoesterase | 6.42e-116 | 46 | 445 | 1 | 347 | Phosphoesterase family. This family includes both bacterial phospholipase C enzymes EC:3.1.4.3, but also eukaryotic acid phosphatases EC:3.1.3.2. |
| cd16013 | AcpA | 1.61e-71 | 44 | 446 | 1 | 367 | acid phosphatase A. Acid phosphatase A catalyzes the hydrolysis reaction via a phosphoseryl intermediate to produce inorganic phosphate and the corresponding alcohol, optimally at low pH. AcpA hydrolyzes a variety of substrates, including p-nitrophenylphosphate (pNPP), p-nitrophenylphosphorylcholine (pNPPC), peptides containing phosphotyrosine, inositol phosphates, AMP, ATP, fructose 1,6-bisphosphate, glucose and fructose 6-phosphates, NADP, and ribose 5-phosphate. AcpA is distinct from histidine ACPs and purple ACPs, as well as class A, B, and C bacterial nonspecific ACPs. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKW17792.1 | 8.34e-161 | 3 | 704 | 10 | 674 |
| ANN20098.1 | 2.09e-147 | 15 | 705 | 10 | 699 |
| QWF84028.1 | 3.33e-145 | 4 | 705 | 3 | 698 |
| SDU45390.1 | 1.35e-144 | 15 | 716 | 10 | 709 |
| QNF45407.1 | 1.90e-144 | 15 | 716 | 10 | 709 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2D1G_A | 1.66e-11 | 36 | 467 | 25 | 470 | ChainA, acid phosphatase [Francisella tularensis subsp. novicida],2D1G_B Chain B, acid phosphatase [Francisella tularensis subsp. novicida] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P15713 | 1.29e-227 | 2 | 705 | 4 | 686 | Non-hemolytic phospholipase C OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=plcN PE=3 SV=2 |
| Q9RGS8 | 5.84e-179 | 2 | 702 | 4 | 695 | Non-hemolytic phospholipase C OS=Burkholderia pseudomallei (strain K96243) OX=272560 GN=plcN PE=1 SV=2 |
| P06200 | 4.95e-138 | 4 | 704 | 9 | 725 | Hemolytic phospholipase C OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=plcH PE=3 SV=2 |
| P9WIA8 | 1.42e-77 | 3 | 470 | 10 | 453 | Phospholipase C D OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=plcD PE=1 SV=1 |
| P0A5R9 | 1.42e-77 | 3 | 470 | 10 | 453 | Phospholipase C D OS=Mycobacterium bovis (strain ATCC BAA-935 / AF2122/97) OX=233413 GN=plcD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
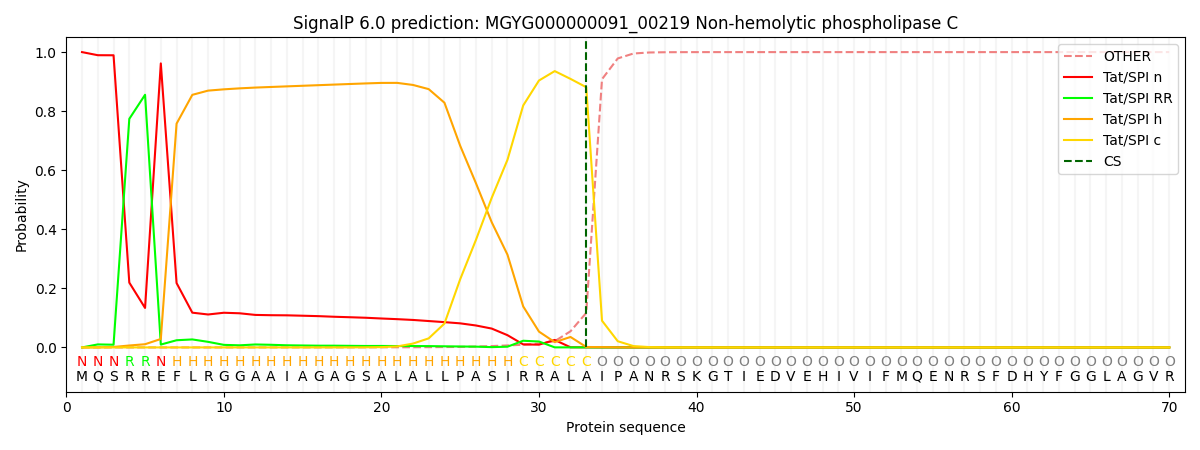
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 0.000000 | 0.999999 | 0.000003 | 0.000000 |