You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000091_05755
You are here: Home > Sequence: MGYG000000091_05755
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Achromobacter xylosoxidans | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Burkholderiales; Burkholderiaceae; Achromobacter; Achromobacter xylosoxidans | |||||||||||
| CAZyme ID | MGYG000000091_05755 | |||||||||||
| CAZy Family | CBM50 | |||||||||||
| CAZyme Description | Murein hydrolase activator NlpD | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 177512; End: 178372 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10871 | nlpD | 3.71e-54 | 11 | 284 | 1 | 317 | murein hydrolase activator NlpD. |
| COG0739 | NlpD | 5.91e-42 | 65 | 286 | 4 | 267 | Murein DD-endopeptidase MepM and murein hydrolase activator NlpD, contain LysM domain [Cell wall/membrane/envelope biogenesis]. |
| pfam01551 | Peptidase_M23 | 1.15e-37 | 187 | 279 | 4 | 96 | Peptidase family M23. Members of this family are zinc metallopeptidases with a range of specificities. The peptidase family M23 is included in this family, these are Gly-Gly endopeptidases. Peptidase family M23 are also endopeptidases. This family also includes some bacterial lipoproteins for which no proteolytic activity has been demonstrated. This family also includes leukocyte cell-derived chemotaxin 2 (LECT2) proteins. LECT2 is a liver-specific protein which is thought to be linked to hepatocyte growth although the exact function of this protein is unknown. |
| COG4942 | EnvC | 1.07e-28 | 171 | 283 | 300 | 418 | Septal ring factor EnvC, activator of murein hydrolases AmiA and AmiB [Cell cycle control, cell division, chromosome partitioning]. |
| cd12797 | M23_peptidase | 1.30e-28 | 187 | 270 | 2 | 85 | M23 family metallopeptidase, also known as beta-lytic metallopeptidase, and similar proteins. This model describes the metallopeptidase M23 family, which includes beta-lytic metallopeptidase and lysostaphin. Members of this family are zinc endopeptidases that lyse bacterial cell wall peptidoglycans; they cleave either the N-acylmuramoyl-Ala bond between the cell wall peptidoglycan and the cross-linking peptide (e.g. beta-lytic endopeptidase) or a bond within the cross-linking peptide (e.g. stapholysin, and lysostaphin). Beta-lytic metallopeptidase, formerly known as beta-lytic protease, has a preference for cleavage of Gly-X bonds and favors hydrophobic or apolar residues on either side. It inhibits growth of sensitive organisms and may potentially serve as an antimicrobial agent. Lysostaphin, produced by Staphylococcus genus, cleaves pentaglycine cross-bridges of cell wall peptidoglycan, acting as autolysins to maintain cell wall metabolism or as toxins and weapons against competing strains. Staphylolysin (also known as LasA) is implicated in a range of processes related to Pseudomonas virulence, including stimulating shedding of the ectodomain of cell surface heparan sulphate proteoglycan syndecan-1, and elastin degradation in connective tissue. Its active site is less constricted and contains a five-coordinate zinc ion with trigonal bipyramidal geometry and two metal-bound water molecules, possibly contributing to its activity against a wider range of substrates than those used by related lytic enzymes, consistent with its multiple roles in Pseudomonas virulence. The family includes members that do not appear to have the conserved zinc-binding site and might be lipoproteins lacking proteolytic activity. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQE58222.1 | 9.73e-194 | 1 | 286 | 1 | 286 |
| CKH36356.1 | 9.73e-194 | 1 | 286 | 1 | 286 |
| QKQ51534.1 | 9.73e-194 | 1 | 286 | 1 | 286 |
| SQG72174.1 | 9.73e-194 | 1 | 286 | 1 | 286 |
| AXA77606.1 | 9.73e-194 | 1 | 286 | 1 | 286 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5J1L_A | 4.92e-15 | 184 | 282 | 63 | 163 | Crystalstructure of Csd1-Csd2 dimer I [Helicobacter pylori 26695],5J1L_C Crystal structure of Csd1-Csd2 dimer I [Helicobacter pylori 26695],5J1M_A Crystal structure of Csd1-Csd2 dimer II [Helicobacter pylori 26695],5J1M_C Crystal structure of Csd1-Csd2 dimer II [Helicobacter pylori 26695] |
| 4BH5_A | 7.48e-14 | 172 | 283 | 23 | 140 | LytMdomain of EnvC, an activator of cell wall amidases in Escherichia coli [Escherichia coli K-12],4BH5_B LytM domain of EnvC, an activator of cell wall amidases in Escherichia coli [Escherichia coli K-12],4BH5_C LytM domain of EnvC, an activator of cell wall amidases in Escherichia coli [Escherichia coli K-12],4BH5_D LytM domain of EnvC, an activator of cell wall amidases in Escherichia coli [Escherichia coli K-12] |
| 6TPI_A | 1.29e-12 | 172 | 283 | 267 | 384 | EnvCbound to the FtsX periplasmic domain [Escherichia coli K-12] |
| 2GU1_A | 2.14e-12 | 177 | 281 | 212 | 327 | Crystalstructure of a zinc containing peptidase from vibrio cholerae [Vibrio cholerae] |
| 5KVP_A | 3.69e-12 | 176 | 270 | 22 | 125 | Solutionstructure of the catalytic domain of zoocin A [Streptococcus equi subsp. zooepidemicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P45682 | 2.25e-50 | 28 | 286 | 28 | 297 | Lipoprotein NlpD/LppB homolog OS=Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) OX=208964 GN=PA3623 PE=3 SV=1 |
| Q46798 | 4.20e-45 | 62 | 286 | 39 | 245 | Uncharacterized lipoprotein YgeR OS=Escherichia coli (strain K12) OX=83333 GN=ygeR PE=3 SV=2 |
| P44833 | 6.48e-43 | 62 | 284 | 144 | 403 | Outer membrane antigenic lipoprotein B OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=lppB PE=3 SV=1 |
| Q56131 | 6.56e-43 | 52 | 286 | 104 | 373 | Murein hydrolase activator NlpD OS=Salmonella typhi OX=90370 GN=nlpD PE=3 SV=2 |
| P40827 | 7.14e-43 | 52 | 286 | 108 | 377 | Murein hydrolase activator NlpD OS=Salmonella typhimurium (strain LT2 / SGSC1412 / ATCC 700720) OX=99287 GN=nlpD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
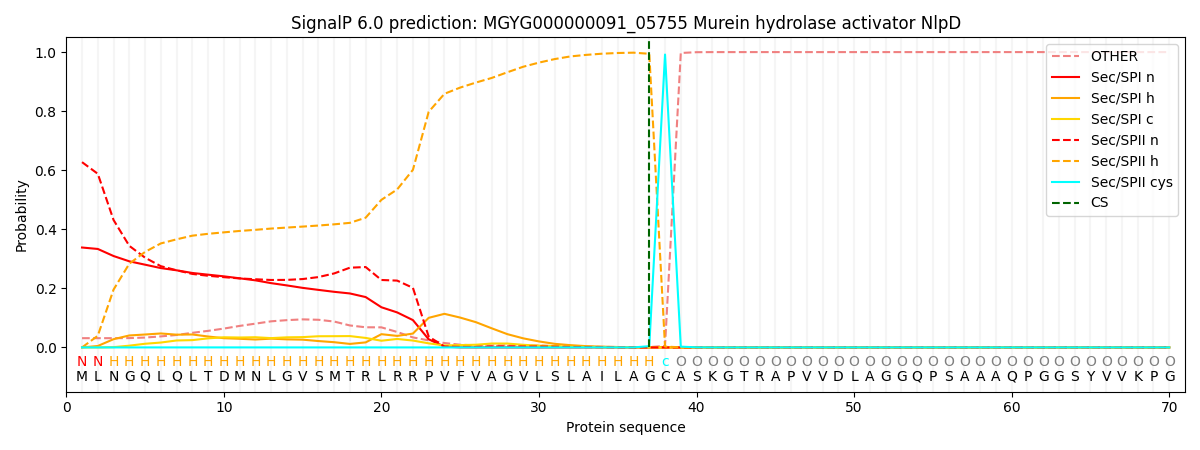
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.008247 | 0.110124 | 0.880867 | 0.000644 | 0.000115 | 0.000034 |
